Species
About ReMap
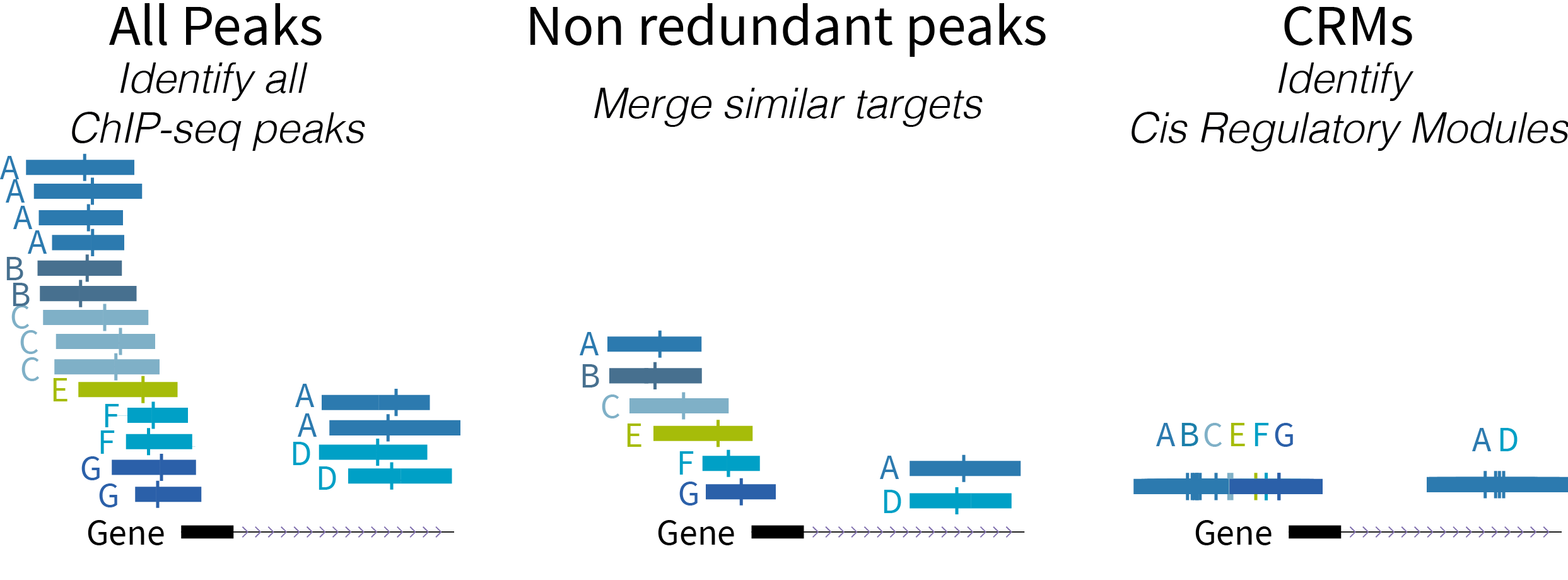
ReMap is an integrative analysis of Homo sapiens and Arabidopsis thaliana transcriptional regulators from DNA-binding experiments such as ChIP-seq, ChIP-exo, DAP-seq from public sources (GEO, ENCODE, ENA). The ReMap human atlas consists of 165 million (M) peaks from 1,135 transcription factors (TFs), transcription coactivators (TCAs) and chromatin-remodeling factors (CRFs). The ReMap Arabidopsis regulatory atlas consists of 2.6M peaks from 372 transcriptionnal regulators. We provide also a catalog of 33 histone modifications and variants across 4.5M peaks. All catalogs are available to browse or download either for a given TF or biotype (cell line, tissue), or for the entire dataset.
ReMap dataset growth
Papers using ReMap
ReMap 2020: a database of regulatory regions from an integrative analysis of Human and Arabidopsis DNA-binding sequencing experiments
Jeanne Chèneby, Zacharie Ménétrier, Martin Mestdagh, Thomas Rosnet, Allyssa Douida, Wassim Rhalloussi, Aurélie Bergon, Fabrice Lopez, Benoit Ballester. Nucleic Acids Research, 29 October 2019, https://doi.org/10.1093/nar/gkz945
Jeanne Chèneby, Zacharie Ménétrier, Martin Mestdagh, Thomas Rosnet, Allyssa Douida, Wassim Rhalloussi, Aurélie Bergon, Fabrice Lopez, Benoit Ballester. Nucleic Acids Research, 29 October 2019, https://doi.org/10.1093/nar/gkz945