Download ReMap
Our data
The ReMap BED files are available to download either for a given transcriptional regulator, by Biotype or for the entire catalog as one very large BED file.
For Homo sapiens the GRCh38/hg38 assembly is currently the supported assembly, but our files can be lifted to hg19 with liftover. We also provide an archive of the ReMap 2018 and 2015 catalogs.
For Arabidopsis thaliana we provide BED files for transcriptional regulators, histones marks, ecotypes and biotype coupled with a given ecotype. The TAIR10 assembly is the only assembly supported by ReMap.
Non-redundant peaks and CRMs annotations are computed across all ReMap datasets and biotypes, thus representing a multi-cellular multi-tissue regulatory map.Data type
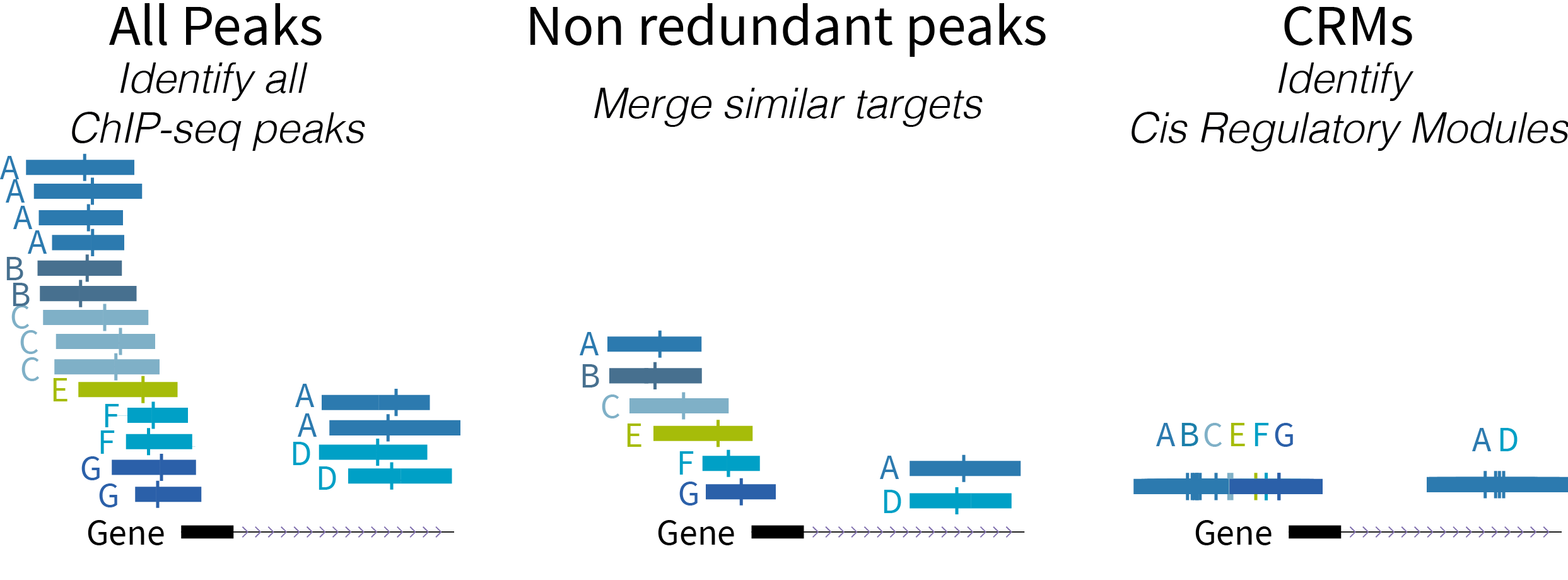
ReMap releases
| Species | All peaks | Merged peaks | CRMs |
|---|---|---|---|
| Homo sapiens | |||
| Arabidopsis thaliana |
| Species | All peaks | Merged peaks | CRMs |
|---|---|---|---|
| Homo sapiens |
| Species | All peaks | Merged peaks |
|---|---|---|
| Homo sapiens |
| Species | atyPeak peaks | About atyPeak |
|---|---|---|
| Homo sapiens |
AtyPeak is a deep-learning based method to identify ChIP-seq peaks that have “atypical profiles” as described in Ferre et al.
Here the analysis was restricted to the densest 65K CRM in the human genome, and to selected TFs and selected datasets. For more information, please see the atyPeak paper in Ferre et al. and at Github. |
ReMap data
| Target name | Ecotype | Biotype | Condition | Source | Species | Experiment | Peaks |
|---|---|---|---|---|---|---|---|
| AATF | LAL-B | GEO | Homo sapiens | GSE93626 | Download (96,653) | ||
| AATF | NALM-6 | GEO | Homo sapiens | GSE93626 | Download (14,991) | ||
| ABF1 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (18,841) |
| ABF1 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (9,819) |
| ABF3 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (21,632) |
| ABF3 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (19,565) |
| ABF4 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (16,755) |
| ABF4 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (12,940) |
| ABI5 | Col-0 | Col-0_seedling | 36h-wt | GEO | Arabidopsis thaliana | GSE60142 | Download (3,323) |
| ABI5 | Col-0 | Col-0_seedling | 36h-ypet | GEO | Arabidopsis thaliana | GSE60142 | Download (8,475) |
| ABI5 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (458) |
| ABR1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (168) |
| ADNP | K-562 | ENCODE | Homo sapiens | ENCSR440VKE | Download (7,766) | ||
| AEBP2 | HEK293 | ENCODE | Homo sapiens | ENCSR769JRS | Download (639) | ||
| AFF1 | K-562 | ENCODE | Homo sapiens | ENCSR241LIH | Download (9,040) | ||
| AFF1 | K-562 | ENCODE | Homo sapiens | ENCSR426URK | Download (25,369) | ||
| AFF1 | MV4-11 | GEO | Homo sapiens | GSE79899 | Download (288) | ||
| AFF1 | L826 | GEO | Homo sapiens | GSE83671 | Download (1,363) | ||
| AFF4 | HCT-116 | SERUM | GEO | Homo sapiens | GSE30267 | Download (1,102) | |
| AFF4 | HCT-116 | STARVED | GEO | Homo sapiens | GSE30267 | Download (394) | |
| AFF4 | HCT-116 | GEO | Homo sapiens | GSE47938 | Download (2,089) | ||
| AFF4 | CD4 | TH1_BAY | GEO | Homo sapiens | GSE62482 | Download (11,253) | |
| AFF4 | CD4 | TH1_DMSO | GEO | Homo sapiens | GSE62482 | Download (18,520) | |
| AFF4 | HEK293T | GEO | Homo sapiens | GSE34097 | Download (266) | ||
| AFF4 | HeLa | GEO | Homo sapiens | GSE40632 | Download (40,361) | ||
| AFF4 | HeLa | DOX | GEO | Homo sapiens | GSE40632 | Download (48,199) | |
| AFF4 | HeLa | DOX_EGF | GEO | Homo sapiens | GSE40632 | Download (46,285) | |
| AFF4 | HeLa | EGF | GEO | Homo sapiens | GSE40632 | Download (42,387) | |
| AG | Ler-0 | Ler-0_inflorescence | 4w | GEO | Arabidopsis thaliana | GSE45938 | Download (3,332) |
| AGL15 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,693) |
| AGL8 | Col-0 | Col-0_Inflorescence-meristem | 6w | GEO | Arabidopsis thaliana | GSE108455 | Download (16,681) |
| AGL8 | Ler | Ler_gynoecium | 6w | GEO | Arabidopsis thaliana | GSE108455 | Download (12,754) |
| AGL8 | Col-0xLer | Col-0xLer_gynoecium-silique-fruit | stage12-16 | GEO | Arabidopsis thaliana | GSE79554 | Download (2,824) |
| AGO1 | Col-0 | Col-0_seedling | 10d-mutant | GEO | Arabidopsis thaliana | GSE95301 | Download (25,646) |
| AGO1 | Col-0 | Col-0_seedling | 10d-wt | GEO | Arabidopsis thaliana | GSE95301 | Download (22,886) |
| AGO1 | Col-0 | Col-0_seedling | 10d-wt-BTH | GEO | Arabidopsis thaliana | GSE95301 | Download (23,267) |
| AGO1 | Col-0 | Col-0_seedling | 10d-wt_BTHcontrol | GEO | Arabidopsis thaliana | GSE95301 | Download (23,986) |
| AGO1 | Col-0 | Col-0_seedling | 10d-wt_Cold | GEO | Arabidopsis thaliana | GSE95301 | Download (20,765) |
| AGO1 | Col-0 | Col-0_seedling | 10d-wt_Coldcontrol | GEO | Arabidopsis thaliana | GSE95301 | Download (17,902) |
| AGO1 | Col-0 | Col-0_seedling | 10d-wt_Flg22 | GEO | Arabidopsis thaliana | GSE95301 | Download (19,145) |
| AGO1 | Col-0 | Col-0_seedling | 10d-wt_Flg22control | GEO | Arabidopsis thaliana | GSE95301 | Download (22,997) |
| AGO1 | Col-0 | Col-0_seedling | 10d-wt_IAA | GEO | Arabidopsis thaliana | GSE95301 | Download (22,524) |
| AGO1 | Col-0 | Col-0_seedling | 10d-wt_IAAcontrol | GEO | Arabidopsis thaliana | GSE95301 | Download (24,522) |
| AGO1 | Col-0 | Col-0_seedling | 10d-wt_MeJA | GEO | Arabidopsis thaliana | GSE95301 | Download (11,007) |
| AGO1 | Col-0 | Col-0_seedling | 10d-wt_MeJAcontrol | GEO | Arabidopsis thaliana | GSE95301 | Download (12,131) |
| AGO1 | K-562 | ENCODE | Homo sapiens | ENCSR641BSL | Download (23,479) | ||
| AGO1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR555ZMV | Download (60,698) | ||
| AGO2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR151NQL | Download (100,658) | ||
| AHR | Hep-G2 | ENCODE | Homo sapiens | ENCSR412ZDC | Download (211,391) | ||
| AHR | GM01310 | 3MC | GEO | Homo sapiens | GSE116632 | Download (25,821) | |
| AHR | GM01310 | DMSO | GEO | Homo sapiens | GSE116632 | Download (27,145) | |
| AHR | MCF-7 | GEO | Homo sapiens | GSE41820 | Download (194,004) | ||
| AHR | MCF-7 | DMSO_1d | GEO | Homo sapiens | GSE90550 | Download (9,903) | |
| AHR | MCF-7 | DMSO_45min | GEO | Homo sapiens | GSE90550 | Download (29,405) | |
| AHR | MCF-7 | TCDD_1d | GEO | Homo sapiens | GSE90550 | Download (10,714) | |
| AHR | MCF-7 | TCDD_45min | GEO | Homo sapiens | GSE90550 | Download (59,381) | |
| AHRR | MCF-7 | DMSO_1d | GEO | Homo sapiens | GSE90550 | Download (3,265) | |
| AIL6 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (249) |
| AIL7 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (170) |
| AIL7 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (2,186) |
| AL5 | Col-0 | Col-0_whole-plant | 3w | GEO | Arabidopsis thaliana | GSE56706 | Download (664) |
| AL5 | Col-0 | Col-0_whole-plant | 3w-mutant | GEO | Arabidopsis thaliana | GSE56706 | Download (308) |
| AMS | Col | Col_cells | GEO | Arabidopsis thaliana | GSE16940 | Download (545) | |
| ANAC016 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (6,873) |
| ANAC020 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,040) |
| ANAC028 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,800) |
| ANAC038 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (710) |
| ANAC050 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (5,207) |
| ANAC050 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (9,484) |
| ANAC057 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,498) |
| ANAC057 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (6,259) |
| ANAC058 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,260) |
| ANAC075 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,205) |
| ANAC076 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (4,672) |
| ANAC094 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (349) |
| ANAC096 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,291) |
| ANAC096 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (1,459) |
| ANAC102 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (14,732) |
| ANAC102 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (12,417) |
| ANAC103 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,548) |
| ANAC103 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (2,678) |
| ANAC13 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,080) |
| ANL2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (536) |
| AP1 | Ler | Ler_inflorescence | 4w-2d | GEO | Arabidopsis thaliana | GSE46986 | Download (2,172) |
| AP1 | Col | Col_apical-meristem | DEX | GEO | Arabidopsis thaliana | GSE20176 | Download (8,289) |
| AP2 | Col-0 | Col-0_inflorescence | GEO | Arabidopsis thaliana | GSE21301 | Download (1,967) | |
| AP3 | Ler | Ler_flower | 4w-dex | GEO | Arabidopsis thaliana | GSE38358 | Download (9,074) |
| APC | HCT-116 | GEO | Homo sapiens | GSE103894 | Download (48,607) | ||
| APRR1 | C24 | C24_seedling | 2w | GEO | Arabidopsis thaliana | GSE35952 | Download (487) |
| APRR5 | Col-0 | Col-0_whole-plant | 18d | GEO | Arabidopsis thaliana | GSE36361 | Download (8,167) |
| APRR7 | Col-2 | Col-2_seedling | 2w | GEO | Arabidopsis thaliana | GSE49282 | Download (4,713) |
| AR | LNCaP-C4-2B | GEO | Homo sapiens | GSE40050 | Download (279) | ||
| AR | LNCaP-C4-2B | DHT | GEO | Homo sapiens | GSE40050 | Download (3,480) | |
| AR | LNCaP-C4-2B | ETOH_SHCTR | GEO | Homo sapiens | GSE61268 | Download (1,097) | |
| AR | LNCaP-C4-2B | R1881_SHCTR | GEO | Homo sapiens | GSE61268 | Download (5,254) | |
| AR | LNCaP-C4-2B | GEO | Homo sapiens | GSE72714 | Download (4,194) | ||
| AR | LNCaP-C4-2B | SR2211 | GEO | Homo sapiens | GSE72714 | Download (9) | |
| AR | MDA-MB-453 | GEO | Homo sapiens | GSE74069 | Download (275) | ||
| AR | MDA-MB-453 | DHT | GEO | Homo sapiens | GSE74069 | Download (21,247) | |
| AR | MDA-MB-453 | MPA | GEO | Homo sapiens | GSE74069 | Download (9,573) | |
| AR | NSC | DHT_10nM | GEO | Homo sapiens | GSE86454 | Download (152) | |
| AR | myofibroblast | GEO | Homo sapiens | GSE90772 | Download (24,722) | ||
| AR | 22Rv1 | GEO | Homo sapiens | GSE96652 | Download (20,740) | ||
| AR | LNCaP | DHT-1nM | GEO | Homo sapiens | GSE94577 | Download (145) | |
| AR | LNCaP | GEO | Homo sapiens | GSE94682 | Download (23,551) | ||
| AR | LNCaP | r1881 | GEO | Homo sapiens | GSE94682 | Download (59,610) | |
| AR | VCaP | GEO | Homo sapiens | GSE98809 | Download (21,896) | ||
| AR | LTAD | DHT-1nM | GEO | Homo sapiens | GSE94577 | Download (801) | |
| AR | LTAD | EtOH | GEO | Homo sapiens | GSE94577 | Download (8,371) | |
| AR | LTAD | siCOBLL1 | GEO | Homo sapiens | GSE94577 | Download (113) | |
| AR | LTAD | siControl | GEO | Homo sapiens | GSE94577 | Download (44,574) | |
| AR | VCaP-LTAD | DHT_10nM | GEO | Homo sapiens | GSE94577 | Download (17,076) | |
| AR | VCaP-LTAD | DHT_1nM | GEO | Homo sapiens | GSE94577 | Download (15,459) | |
| AR | LNCaP-C4-2 | GEO | Homo sapiens | GSE97831 | Download (191) | ||
| AR | MCF-7 | ENA | Homo sapiens | ERP001226 | Download (58,568) | ||
| AR | MCF-7 | GEO | Homo sapiens | GSE104399 | Download (1,119) | ||
| AR | MDA-MB-453 | ENA | Homo sapiens | ERP001226 | Download (23,406) | ||
| AR | MDA-MB-453 | SHFOXA1 | ENA | Homo sapiens | ERP003272 | Download (179,906) | |
| AR | MDA-MB-453 | ENA | Homo sapiens | ERP003503 | Download (72,884) | ||
| AR | MDA-MB-453 | FOXA1 | ENA | Homo sapiens | ERP003503 | Download (74,086) | |
| AR | epididymis | HEE | GEO | Homo sapiens | GSE109061 | Download (153) | |
| AR | epididymis | HEE_R1881 | GEO | Homo sapiens | GSE109061 | Download (12,349) | |
| AR | 22Rv1 | GEO | Homo sapiens | GSE40050 | Download (77) | ||
| AR | 22Rv1 | DHT | GEO | Homo sapiens | GSE40050 | Download (314) | |
| AR | 22Rv1 | R1881 | GEO | Homo sapiens | GSE80742 | Download (27,025) | |
| AR | 22Rv1 | siARFL | GEO | Homo sapiens | GSE80742 | Download (2,407) | |
| AR | 22Rv1 | siARFL_R1881 | GEO | Homo sapiens | GSE80742 | Download (24,265) | |
| AR | 22Rv1 | GEO | Homo sapiens | GSE85558 | Download (8,325) | ||
| AR | 22Rv1 | Dox | GEO | Homo sapiens | GSE85558 | Download (1,364) | |
| AR | DU145 | GEO | Homo sapiens | GSE47987 | Download (13,396) | ||
| AR | DU145 | ARC562S | GEO | Homo sapiens | GSE47987 | Download (22) | |
| AR | DU145 | ARG56W | GEO | Homo sapiens | GSE47987 | Download (202) | |
| AR | DU145 | ARQ6540X | GEO | Homo sapiens | GSE47987 | Download (18,791) | |
| AR | DU145 | FOXA1 | GEO | Homo sapiens | GSE47987 | Download (11,651) | |
| AR | DU145 | FOXA1_ARC562S | GEO | Homo sapiens | GSE47987 | Download (14) | |
| AR | DU145 | FOXA1_ARG56W | GEO | Homo sapiens | GSE47987 | Download (2) | |
| AR | DU145 | FOXA1_ARQ6540X | GEO | Homo sapiens | GSE47987 | Download (24,287) | |
| AR | DU145 | FOXA1_HIGH | GEO | Homo sapiens | GSE47987 | Download (747) | |
| AR | DUCAP | GEO | Homo sapiens | GSE70679 | Download (7) | ||
| AR | DUCAP | ANDROGEN | GEO | Homo sapiens | GSE70679 | Download (24,610) | |
| AR | LHSAR | HOXB13 | GEO | Homo sapiens | GSE56288 | Download (28,987) | |
| AR | LHSAR | LACZ | GEO | Homo sapiens | GSE56288 | Download (156,321) | |
| AR | LNCaP | GEO | Homo sapiens | GSE110655 | Download (72,127) | ||
| AR | LNCaP | ERG | GEO | Homo sapiens | GSE110655 | Download (80,270) | |
| AR | LNCaP | 1F5 | GEO | Homo sapiens | GSE30623 | Download (18,917) | |
| AR | LNCaP | 1F5_SIFOXA1 | GEO | Homo sapiens | GSE30623 | Download (28,929) | |
| AR | LNCaP | GEO | Homo sapiens | GSE31294 | Download (2,174) | ||
| AR | LNCaP | KDP53 | GEO | Homo sapiens | GSE31294 | Download (43) | |
| AR | LNCaP | SHCTR_R1881 | GEO | Homo sapiens | GSE37345 | Download (9,068) | |
| AR | LNCaP | SHCTR_R1881_HD | GEO | Homo sapiens | GSE37345 | Download (20) | |
| AR | LNCaP | SHFOXA1_R1881 | GEO | Homo sapiens | GSE37345 | Download (29,635) | |
| AR | LNCaP | SHFOXA1_R1881_HD | GEO | Homo sapiens | GSE37345 | Download (1,077) | |
| AR | LNCaP | DHT | GEO | Homo sapiens | GSE40050 | Download (1,851) | |
| AR | LNCaP | GEO | Homo sapiens | GSE43720 | Download (16,706) | ||
| AR | LNCaP | DHT | GEO | Homo sapiens | GSE43720 | Download (38,962) | |
| AR | LNCaP | SHCTR | GEO | Homo sapiens | GSE47120 | Download (1,484) | |
| AR | LNCaP | SHETV1 | GEO | Homo sapiens | GSE47120 | Download (7) | |
| AR | LNCaP | SHCTR | GEO | Homo sapiens | GSE52725 | Download (84,761) | |
| AR | LNCaP | SHGATA2 | GEO | Homo sapiens | GSE52725 | Download (9,525) | |
| AR | LNCaP | DHT24H | GEO | Homo sapiens | GSE58428 | Download (22,509) | |
| AR | LNCaP | DHT24H_SHFOXA1 | GEO | Homo sapiens | GSE58428 | Download (934) | |
| AR | LNCaP | ETOH_SHCTR | GEO | Homo sapiens | GSE61268 | Download (768) | |
| AR | LNCaP | HOTAIR | GEO | Homo sapiens | GSE61268 | Download (5,115) | |
| AR | LNCaP | R1881 | GEO | Homo sapiens | GSE61268 | Download (6,138) | |
| AR | LNCaP | R1881_HOTAIR | GEO | Homo sapiens | GSE61268 | Download (15,098) | |
| AR | LNCaP | R1881_SHCTR | GEO | Homo sapiens | GSE61268 | Download (1,350) | |
| AR | LNCaP | GEO | Homo sapiens | GSE62492 | Download (1,219) | ||
| AR | LNCaP | R1881 | GEO | Homo sapiens | GSE62492 | Download (25,773) | |
| AR | LNCaP | SHCTR_DHT | GEO | Homo sapiens | GSE62492 | Download (52,727) | |
| AR | LNCaP | SHFOXP1_DHT | GEO | Homo sapiens | GSE62492 | Download (47,706) | |
| AR | LNCaP | GEO | Homo sapiens | GSE63202 | Download (13,285) | ||
| AR | LNCaP | GEO | Homo sapiens | GSE64656 | Download (15,308) | ||
| AR | LNCaP | ETOH | GEO | Homo sapiens | GSE69043 | Download (21,094) | |
| AR | LNCaP | PHFRPMIFCS | GEO | Homo sapiens | GSE69043 | Download (20) | |
| AR | LNCaP | R1881 | GEO | Homo sapiens | GSE69043 | Download (29,351) | |
| AR | LNCaP | RPMIFBS | GEO | Homo sapiens | GSE69043 | Download (5,714) | |
| AR | LNCaP | SHFOXA1_PHFRPMIFCS | GEO | Homo sapiens | GSE69043 | Download (1,075) | |
| AR | LNCaP | SHFOXA1_RPMIFBS | GEO | Homo sapiens | GSE69043 | Download (2,911) | |
| AR | LNCaP | SHGATA2_ETOH | GEO | Homo sapiens | GSE69043 | Download (16,306) | |
| AR | LNCaP | SHGATA2_R1881 | GEO | Homo sapiens | GSE69043 | Download (47,489) | |
| AR | LNCaP | GEO | Homo sapiens | GSE79357 | Download (2,371) | ||
| AR | LNCaP | NOV | GEO | Homo sapiens | GSE79357 | Download (1,560) | |
| AR | LNCaP | GEO | Homo sapiens | GSE80256 | Download (62,064) | ||
| AR | LNCaP | DHT | GEO | Homo sapiens | GSE83860 | Download (38,177) | |
| AR | LNCaP | DHT_TNFA | GEO | Homo sapiens | GSE83860 | Download (31,301) | |
| AR | LNCaP | DMSO | GEO | Homo sapiens | GSE83860 | Download (1,157) | |
| AR | LNCaP | GEO | Homo sapiens | GSE85558 | Download (29,939) | ||
| AR | LNCaP | HNF4G_ovexp | GEO | Homo sapiens | GSE85558 | Download (13,422) | |
| AR | LNCaP | 3279 | GEO | Homo sapiens | GSE86456 | Download (123) | |
| AR | LNCaP | DHT_100nM_3279 | GEO | Homo sapiens | GSE86456 | Download (1,174) | |
| AR | LNCaP | DHT_100nM_N20 | GEO | Homo sapiens | GSE86456 | Download (1,658) | |
| AR | LNCaP | N20 | GEO | Homo sapiens | GSE86456 | Download (129) | |
| AR | LNCaP | Bag1L_KO_Bag1L_Rescue_DHT_4h | GEO | Homo sapiens | GSE89938 | Download (24,109) | |
| AR | LNCaP | Bag-1L_KO_DHT | GEO | Homo sapiens | GSE89938 | Download (15,111) | |
| AR | LNCaP | Bag-1L_KO_Veh | GEO | Homo sapiens | GSE89938 | Download (10,154) | |
| AR | LNCaP | Bag-1L_WT | GEO | Homo sapiens | GSE89938 | Download (6,996) | |
| AR | LNCaP | Bag-1L_WT_DHT | GEO | Homo sapiens | GSE89938 | Download (31,045) | |
| AR | LNCaP | DHT_Bag-1L-CMut | GEO | Homo sapiens | GSE89938 | Download (19,707) | |
| AR | LNCaP | Talen_DHT | GEO | Homo sapiens | GSE89938 | Download (47,794) | |
| AR | LNCaP | Talen_Veh | GEO | Homo sapiens | GSE89938 | Download (6,544) | |
| AR | LNCaP | DHT | GEO | Homo sapiens | GSE92347 | Download (8,451) | |
| AR | LNCaP | SRC_DHT | GEO | Homo sapiens | GSE92347 | Download (2,574) | |
| AR | PLHSAR | FOXA1_HOXB13 | GEO | Homo sapiens | GSE56288 | Download (230,564) | |
| AR | prostate | GEO | Homo sapiens | GSE56288 | Download (68,830) | ||
| AR | prostate | cancer | GEO | Homo sapiens | GSE56288 | Download (162,232) | |
| AR | prostate | DHT | GEO | Homo sapiens | GSE61838 | Download (20,568) | |
| AR | prostate | ETOH | GEO | Homo sapiens | GSE61838 | Download (3) | |
| AR | prostate | GEO | Homo sapiens | GSE65478 | Download (19,899) | ||
| AR | prostate | cancer-associated-fibroblasts | GEO | Homo sapiens | GSE90772 | Download (37,526) | |
| AR | prostate | fetal-fibroblast | GEO | Homo sapiens | GSE90772 | Download (76,745) | |
| AR | VCaP | shCt | GEO | Homo sapiens | GSE110655 | Download (24,132) | |
| AR | VCaP | shERG | GEO | Homo sapiens | GSE110655 | Download (55,093) | |
| AR | VCaP | GEO | Homo sapiens | GSE28950 | Download (4,862) | ||
| AR | VCaP | DHAT_18H | GEO | Homo sapiens | GSE28950 | Download (36,524) | |
| AR | VCaP | DHAT_2H | GEO | Homo sapiens | GSE28950 | Download (63,320) | |
| AR | VCaP | GEO | Homo sapiens | GSE32892 | Download (19,852) | ||
| AR | VCaP | R1881 | GEO | Homo sapiens | GSE32892 | Download (65,624) | |
| AR | VCaP | R1881_10C26 | GEO | Homo sapiens | GSE32892 | Download (27,397) | |
| AR | VCaP | R1881_10C30 | GEO | Homo sapiens | GSE32892 | Download (16,099) | |
| AR | VCaP | R1881_1C26 | GEO | Homo sapiens | GSE32892 | Download (24,418) | |
| AR | VCaP | R1881_1C30 | GEO | Homo sapiens | GSE32892 | Download (34,074) | |
| AR | VCaP | GEO | Homo sapiens | GSE56086 | Download (122,097) | ||
| AR | VCaP | SHPIAS1_R1881 | GEO | Homo sapiens | GSE56086 | Download (111,464) | |
| AR | VCaP | DHT24H | GEO | Homo sapiens | GSE58428 | Download (50,287) | |
| AR | VCaP | DHT24H_SHFOXA1 | GEO | Homo sapiens | GSE58428 | Download (40,539) | |
| AR | VCaP | DHT24H_SHFOXP1 | GEO | Homo sapiens | GSE58428 | Download (69,103) | |
| AR | VCaP | DHT24H_SHRUNX1 | GEO | Homo sapiens | GSE58428 | Download (42,569) | |
| AR | VCaP | ETOH24H | GEO | Homo sapiens | GSE58428 | Download (1,427) | |
| AR | VCaP | DHT | GEO | Homo sapiens | GSE79128 | Download (25,392) | |
| AR | VCaP | R1881 | GEO | Homo sapiens | GSE79128 | Download (11,265) | |
| AR | VCaP | SH1_DHT | GEO | Homo sapiens | GSE79128 | Download (34,691) | |
| AR | VCaP | SH1_R1881 | GEO | Homo sapiens | GSE79128 | Download (27,676) | |
| AR | VCaP | SH2_DHT | GEO | Homo sapiens | GSE79128 | Download (28,035) | |
| AR | VCaP | SH2_R1881 | GEO | Homo sapiens | GSE79128 | Download (29,245) | |
| AR | VCaP | SH3_DHT | GEO | Homo sapiens | GSE79128 | Download (18,718) | |
| AR | VCaP | R1881_30M | GEO | Homo sapiens | GSE84432 | Download (71,965) | |
| AR | VCaP | R1881_4H | GEO | Homo sapiens | GSE84432 | Download (73,189) | |
| AR | VCaP | GEO | Homo sapiens | GSE92347 | Download (889) | ||
| AR | VCaP | DHT | GEO | Homo sapiens | GSE92347 | Download (1,828) | |
| AR | A-375 | GEO | Homo sapiens | GSE116189 | Download (7,228) | ||
| AR | A-375 | SLNCR | GEO | Homo sapiens | GSE116189 | Download (9,813) | |
| AR | LNCaP-abl | GEO | Homo sapiens | GSE39459 | Download (69,336) | ||
| AR | LNCaP-abl | DMSO | GEO | Homo sapiens | GSE80238 | Download (30,056) | |
| AR | MCF-7 | GEO | Homo sapiens | GSE48930 | Download (60,294) | ||
| AR | MDA-MB-453 | R1881 | GEO | Homo sapiens | GSE70161 | Download (793) | |
| AR | MDA-MB-453 | R1881_SICTR | GEO | Homo sapiens | GSE70161 | Download (32,838) | |
| AR | MDA-MB-453 | R1881_SIPIAS1 | GEO | Homo sapiens | GSE70161 | Download (44,998) | |
| AR | PC-3 | GEO | Homo sapiens | GSE54110 | Download (2,241) | ||
| AR | PC-3 | R1881 | GEO | Homo sapiens | GSE54110 | Download (67,317) | |
| AR | breast | tumor_Female_8 | GEO | Homo sapiens | GSE104399 | Download (4,186) | |
| AR | breast | tumor_Male_10 | GEO | Homo sapiens | GSE104399 | Download (33,018) | |
| AR | breast | tumor_Male_17 | GEO | Homo sapiens | GSE104399 | Download (2,631) | |
| AR | breast | tumor_Male_18 | GEO | Homo sapiens | GSE104399 | Download (9,017) | |
| AR | breast | tumor_Male_20 | GEO | Homo sapiens | GSE104399 | Download (13,466) | |
| AR | breast | tumor_Male_26 | GEO | Homo sapiens | GSE104399 | Download (34,301) | |
| AR | breast | tumor_Male_28 | GEO | Homo sapiens | GSE104399 | Download (11,384) | |
| AR | breast | tumor_Male_30 | GEO | Homo sapiens | GSE104399 | Download (5,579) | |
| AR | breast | tumor_Male_7 | GEO | Homo sapiens | GSE104399 | Download (5,983) | |
| AR | breast | tumor_Male_8 | GEO | Homo sapiens | GSE104399 | Download (7,896) | |
| AR | LNCaP | ENA | Homo sapiens | ERP001226 | Download (46,671) | ||
| AR | LNCaP | ENA | Homo sapiens | ERP003503 | Download (40,165) | ||
| AR | LNCaP | FOXA1 | ENA | Homo sapiens | ERP003503 | Download (76,135) | |
| ARF | Col-0 | Col-0_seedling-etiolated | 5d | GEO | Arabidopsis thaliana | GSE51770 | Download (2,567) |
| ARHGAP35 | K-562 | ENCODE | Homo sapiens | ENCSR571BUF | Download (5,484) | ||
| ARID1A | LNCaP | GEO | Homo sapiens | GSE94682 | Download (13,289) | ||
| ARID1A | LNCaP | r1881 | GEO | Homo sapiens | GSE94682 | Download (28,266) | |
| ARID1A | RMG-I | GEO | Homo sapiens | GSE104545 | Download (47,140) | ||
| ARID1A | Hep-G2 | GEO | Homo sapiens | GSE69566 | Download (17,397) | ||
| ARID1B | Hep-G2 | GEO | Homo sapiens | GSE69566 | Download (14,167) | ||
| ARID1B | K-562 | ENCODE | Homo sapiens | ENCSR822CCM | Download (70,488) | ||
| ARID2 | Aska-SS | GEO | Homo sapiens | GSE108025 | Download (34,024) | ||
| ARID2 | Aska-SS | shSSX | GEO | Homo sapiens | GSE108025 | Download (45,178) | |
| ARID2 | K-562 | ENCODE | Homo sapiens | ENCSR491EBY | Download (9,889) | ||
| ARID2 | Hep-G2 | GEO | Homo sapiens | GSE69566 | Download (21,533) | ||
| ARID3A | MCF-7 | ENCODE | Homo sapiens | ENCSR231ENE | Download (1,360) | ||
| ARID3A | Hep-G2 | GEO | Homo sapiens | GSE97661 | Download (8,850) | ||
| ARID3A | GM12878 | ENCODE | Homo sapiens | ENCSR725LYT | Download (22,263) | ||
| ARID3A | GM12878 | ENCODE | Homo sapiens | ENCSR778UBR | Download (17,125) | ||
| ARID3A | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EDP | Download (42,797) | ||
| ARID3A | K-562 | ENCODE | Homo sapiens | ENCSR000EFY | Download (22,191) | ||
| ARID4B | Hep-G2 | ENCODE | Homo sapiens | ENCSR988LZG | Download (94,553) | ||
| ARID5B | Jurkat | GEO | Homo sapiens | GSE97512 | Download (17,087) | ||
| ARID6 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,143) |
| ARNT | 786-M1A | GEO | Homo sapiens | GSE98012 | Download (445) | ||
| ARNT | K-562 | ENCODE | Homo sapiens | ENCSR613NUC | Download (26,693) | ||
| ARNT | K-562 | ENCODE | Homo sapiens | ENCSR669NFS | Download (2,130) | ||
| ARNT | RCC10 | GEO | Homo sapiens | GSE101063 | Download (1,582) | ||
| ARNT | HUVEC-C | GEO | Homo sapiens | GSE89836 | Download (543) | ||
| ARNT | MCF-7 | GEO | Homo sapiens | GSE41820 | Download (369,183) | ||
| ARNT | T-47D | HAEGIN2 | GEO | Homo sapiens | GSE59935 | Download (135,183) | |
| ARNT | GM12878 | ENCODE | Homo sapiens | ENCSR590KEQ | Download (15,548) | ||
| ARNT | HEK293T | ENCODE | Homo sapiens | ENCSR760UKJ | Download (1,501) | ||
| ARNT | Hep-G2 | ENCODE | Homo sapiens | ENCSR029IBC | Download (1,150) | ||
| ARNT | K-562 | ENCODE | Homo sapiens | ENCSR155KHM | Download (12,328) | ||
| ARNTL | U2OS | GEO | Homo sapiens | GSE44236 | Download (20,741) | ||
| ARNTL | U2OS | DMOG | GEO | Homo sapiens | GSE85096 | Download (32,023) | |
| ARNTL | U2OS | DMSO | GEO | Homo sapiens | GSE85096 | Download (12,215) | |
| ARNTL | U2OS | trough_DMOG | GEO | Homo sapiens | GSE85096 | Download (17,969) | |
| ARNTL | U2OS | trough_DMSO | GEO | Homo sapiens | GSE85096 | Download (8,090) | |
| ARR1 | Col-0 | Col-0_seedling | 3d-6BA-4h | GEO | Arabidopsis thaliana | GSE94486 | Download (4,328) |
| ARR1 | Col-0 | Col-0_seedling | 3d-6BA-72h | GEO | Arabidopsis thaliana | GSE94486 | Download (21,115) |
| ARR1 | Col-0 | Col-0_seedling | 3d-Dex-4h | GEO | Arabidopsis thaliana | GSE94486 | Download (12,934) |
| ARR1 | Col-0 | Col-0_seedling | 3d-mock-4h-etiolated | GEO | Arabidopsis thaliana | GSE94486 | Download (3,203) |
| ARR1 | Col-0 | Col-0_seedling | 3d-mock-4h-LD | GEO | Arabidopsis thaliana | GSE94486 | Download (7,096) |
| ARR10 | Col-0 | Col-0_seedling | 3d-6BA-4h | GEO | Arabidopsis thaliana | GSE94486 | Download (9,700) |
| ARR10 | Col-0 | Col-0_seedling | 3d-mock-4h | GEO | Arabidopsis thaliana | GSE94486 | Download (11,167) |
| ARR12 | Col-0 | Col-0_seedling | 3d-6BA-4h | GEO | Arabidopsis thaliana | GSE94486 | Download (8,438) |
| ARR12 | Col-0 | Col-0_seedling | 3d-mock-4h | GEO | Arabidopsis thaliana | GSE94486 | Download (16,320) |
| ARR14 | Col-0 | Col-0_seedling | 3d-6BA-72h | GEO | Arabidopsis thaliana | GSE94486 | Download (20,651) |
| ARRB1 | LNCaP-C4-2 | GEO | Homo sapiens | GSE55615 | Download (8,363) | ||
| ARRB1 | prostate | GEO | Homo sapiens | GSE55615 | Download (16,464) | ||
| AS2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (132) |
| ASCL1 | G523NS | Dox | GEO | Homo sapiens | GSE87618 | Download (1,553) | |
| ASCL1 | SCLC | ASCLM | GEO | Homo sapiens | GSE61197 | Download (203) | |
| ASCL1 | SCLC | ASCLP | GEO | Homo sapiens | GSE61197 | Download (31,038) | |
| ASCL1 | SCLC | ASCLP_NE | GEO | Homo sapiens | GSE61197 | Download (9,834) | |
| ASCL1 | NCI-H2107 | GEO | Homo sapiens | GSE69394 | Download (22,341) | ||
| ASCL1 | NCI-H82 | GEO | Homo sapiens | GSE69394 | Download (26,105) | ||
| ASCL1 | NCI-H889 | GEO | Homo sapiens | GSE69394 | Download (31,933) | ||
| ASCL1 | H128 | GEO | Homo sapiens | GSE69394 | Download (6,059) | ||
| ASH1L | K-562 | ENCODE | Homo sapiens | ENCSR115BBC | Download (4,958) | ||
| ASH2L | GM12878 | ENCODE | Homo sapiens | ENCSR849WCQ | Download (34,589) | ||
| ASH2L | Hep-G2 | ENCODE | Homo sapiens | ENCSR508LMH | Download (54,945) | ||
| ASH2L | VCaP | GEO | Homo sapiens | GSE60841 | Download (30,838) | ||
| ASH2L | VCaP | R1881 | GEO | Homo sapiens | GSE60841 | Download (40,248) | |
| ASH2L | WA01 | ENCODE | Homo sapiens | ENCSR850KIP | Download (57,856) | ||
| ASIL2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,406) |
| ASXL1 | HEK293T | GEO | Homo sapiens | GSE51673 | Download (9,359) | ||
| AT-GTL1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,356) |
| AT1G14580 | Col-0 | Col-0_roots | GEO | Arabidopsis thaliana | GSE60011 | Download (5,270) | |
| AT1G19000 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,701) |
| AT1G24250 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (451) |
| AT1G49010 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (783) |
| AT1G72740 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (466) |
| AT1G74840 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,790) |
| AT2G15740 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (410) |
| AT2G20110 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,045) |
| AT2G20110 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (23,825) |
| AT2G20400 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (125) |
| AT2G33710 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,013) |
| AT2G40260 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (810) |
| AT3G10030 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (511) |
| AT3G10030 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (1,873) |
| AT3G11100 | Col-0 | Col-0_seedling | 3d-air | GEO | Arabidopsis thaliana | GSE97288 | Download (5,881) |
| AT3G11100 | Col-0 | Col-0_seedling | 3d-C2H4 | GEO | Arabidopsis thaliana | GSE97288 | Download (4,881) |
| AT3G11280 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (897) |
| AT3G42860 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (799) |
| AT3G49930 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,457) |
| AT4G16150 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (661) |
| AT4G33280 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (133) |
| AT5G04390 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (200) |
| AT5G04760 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (22,584) |
| AT5G04760 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (21,019) |
| AT5G05550 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,043) |
| AT5G05550 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (1,860) |
| AT5G08750 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (436) |
| AT5G23930 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,585) |
| AT5G47660 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (13,710) |
| AT5G50360 | Col-0 | Col-0_seedling | 4d-DEX-6h | GEO | Arabidopsis thaliana | GSE80566 | Download (18,922) |
| AT5G56840 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (595) |
| AT5G58900 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (22,621) |
| AT5G60130 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (131) |
| AT5G61620 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,570) |
| ATBZIP42 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (112) |
| ATF1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR253OON | Download (2,278) | ||
| ATF1 | K-562 | ENCODE | Homo sapiens | ENCSR000DNZ | Download (9,811) | ||
| ATF1 | K-562 | ENCODE | Homo sapiens | ENCSR091GVJ | Download (43,543) | ||
| ATF1 | K-562 | ENCODE | Homo sapiens | ENCSR159OCC | Download (3,544) | ||
| ATF2 | GM12878 | ENCODE | Homo sapiens | ENCSR000BQK | Download (33,025) | ||
| ATF2 | GM12878 | ENCODE | Homo sapiens | ENCSR961PPA | Download (18,713) | ||
| ATF2 | HEK293 | ENCODE | Homo sapiens | ENCSR217HTK | Download (34,000) | ||
| ATF2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR047BUZ | Download (70,181) | ||
| ATF2 | K-562 | ENCODE | Homo sapiens | ENCSR869IUD | Download (51,764) | ||
| ATF2 | WA01 | ENCODE | Homo sapiens | ENCSR000BQU | Download (43,936) | ||
| ATF2 | macrophage | GEO | Homo sapiens | GSE80727 | Download (7,829) | ||
| ATF3 | GM00739 | GEO | Homo sapiens | GSE87562 | Download (119,883) | ||
| ATF3 | GM00739 | CSBwt_8h | GEO | Homo sapiens | GSE87562 | Download (100,720) | |
| ATF3 | GM12878 | ENCODE | Homo sapiens | ENCSR000BJY | Download (4,002) | ||
| ATF3 | liver | ENCODE | Homo sapiens | ENCSR205FOW | Download (19,361) | ||
| ATF3 | liver | ENCODE | Homo sapiens | ENCSR480LIS | Download (32,423) | ||
| ATF3 | A-549 | ENCODE | Homo sapiens | ENCSR000BPS | Download (19,201) | ||
| ATF3 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BKE | Download (3,811) | ||
| ATF3 | K-562 | ENCODE | Homo sapiens | ENCSR000BNU | Download (19,250) | ||
| ATF3 | K-562 | ENCODE | Homo sapiens | ENCSR000DOG | Download (866) | ||
| ATF3 | K-562 | ENCODE | Homo sapiens | ENCSR028UIU | Download (79,031) | ||
| ATF3 | HCT-116 | CAMP | GEO | Homo sapiens | GSE74355 | Download (74,099) | |
| ATF3 | HCT-116 | DMSO | GEO | Homo sapiens | GSE74355 | Download (35,240) | |
| ATF3 | HCT-116 | DMSO_KOATF3 | GEO | Homo sapiens | GSE74355 | Download (995) | |
| ATF3 | primary-dermal-fibroblasts | overexpressed | GEO | Homo sapiens | GSE81403 | Download (16,789) | |
| ATF3 | K-562 | ENCODE | Homo sapiens | ENCSR632DCH | Download (26,041) | ||
| ATF3 | WA01 | ENCODE | Homo sapiens | ENCSR000BKC | Download (6,923) | ||
| ATF3 | HCT-116 | ENCODE | Homo sapiens | ENCSR000BUG | Download (2,977) | ||
| ATF4 | Hep-G2 | ENCODE | Homo sapiens | ENCSR669LCD | Download (1,409) | ||
| ATF4 | K-562 | ENCODE | Homo sapiens | ENCSR145TSJ | Download (54,286) | ||
| ATF4 | hMSC | GEO | Homo sapiens | GSE68864 | Download (648) | ||
| ATF4 | DLD-1 | MYC-activated | GEO | Homo sapiens | GSE117240 | Download (222) | |
| ATF7 | GM12878 | ENCODE | Homo sapiens | ENCSR014YCR | Download (69,629) | ||
| ATF7 | Hep-G2 | ENCODE | Homo sapiens | ENCSR545FXC | Download (45,643) | ||
| ATF7 | K-562 | ENCODE | Homo sapiens | ENCSR972ZBV | Download (49,409) | ||
| ATF7 | MCF-7 | ENCODE | Homo sapiens | ENCSR866QPZ | Download (30,318) | ||
| ATHB-15 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (391) |
| ATHB-23 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,057) |
| ATHB-5 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (20,564) |
| ATHB-5 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (20,711) |
| ATHB-5 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (328) |
| ATHB-51 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (126) |
| ATHB-53 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,712) |
| ATHB-6 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (20,225) |
| ATHB-6 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (13,217) |
| ATHB-7 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (23,413) |
| ATHB-7 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (15,259) |
| ATHB-9 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,473) |
| ATHB-X | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (401) |
| ATM | Hep-G2 | ENCODE | Homo sapiens | ENCSR859JGF | Download (2,146) | ||
| ATMYB119 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,117) |
| ATMYB119 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (10,220) |
| ATMYB13 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,272) |
| ATMYB63 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (160) |
| ATRX | LAN6 | KOATRX | GEO | Homo sapiens | GSE70920 | Download (208) | |
| ATRX | K-562 | KOATRX | GEO | Homo sapiens | GSE70920 | Download (193) | |
| ATRX | erythroid | GEO | Homo sapiens | GSE22162 | Download (1,283) | ||
| AZF1 | Col | Col_whole-plant | 30h | GEO | Arabidopsis thaliana | GSE84483 | Download (4,493) |
| AtGRF6 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,289) |
| AtMYB70 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,815) |
| AtMYB81 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (638) |
| AtMYB93 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,397) |
| AtbZIP3 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,458) |
| AtbZIP48 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (206) |
| AtbZIP48 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (4,355) |
| AtbZIP52 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (6,560) |
| BACH1 | K-562 | ENCODE | Homo sapiens | ENCSR740NPG | Download (8,541) | ||
| BACH1 | WA01 | ENCODE | Homo sapiens | ENCSR000EBQ | Download (20,092) | ||
| BACH1 | GM12878 | ENCODE | Homo sapiens | ENCSR585CVE | Download (3,052) | ||
| BACH1 | GM12878 | ENCODE | Homo sapiens | ENCSR636MKU | Download (29,168) | ||
| BACH1 | A-549 | ENCODE | Homo sapiens | ENCSR043EHG | Download (5,895) | ||
| BACH1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR699TNT | Download (9,031) | ||
| BACH1 | K-562 | ENCODE | Homo sapiens | ENCSR000EGD | Download (3,908) | ||
| BACH2 | B-cell | IL2 | GEO | Homo sapiens | GSE102460 | Download (14,896) | |
| BACH2 | B-cell | siBACH2 | GEO | Homo sapiens | GSE102460 | Download (2,432) | |
| BACH2 | DOHH2 | GEO | Homo sapiens | GSE69558 | Download (5,363) | ||
| BACH2 | OCI-Ly7 | GEO | Homo sapiens | GSE44420 | Download (8,814) | ||
| BACH2 | OCI-Ly7 | GEO | Homo sapiens | GSE69558 | Download (2,969) | ||
| BAF155 | LNCaP | GEO | Homo sapiens | GSE110655 | Download (99,727) | ||
| BAF155 | LNCaP | ERG | GEO | Homo sapiens | GSE110655 | Download (113,997) | |
| BAF155 | LNCaP | ERG-R367K | GEO | Homo sapiens | GSE110655 | Download (105,914) | |
| BAF155 | VCaP | shARID1A | GEO | Homo sapiens | GSE110655 | Download (71,543) | |
| BAF155 | VCaP | shCt | GEO | Homo sapiens | GSE110655 | Download (94,888) | |
| BAF155 | VCaP | shERG | GEO | Homo sapiens | GSE110655 | Download (77,256) | |
| BAHD1 | HEK293 | GEO | Homo sapiens | GSE53372 | Download (8,043) | ||
| BAP1 | UM-RC-6 | GEO | Homo sapiens | GSE101987 | Download (1,702) | ||
| BAP1 | UM-RC-6 | BAP1-C91A-mutant | GEO | Homo sapiens | GSE101987 | Download (1,647) | |
| BAP1 | UM-RC-6 | empty-vector | GEO | Homo sapiens | GSE101987 | Download (1,460) | |
| BATF | OCI-Ly10 | GEO | Homo sapiens | GSE56857 | Download (10,240) | ||
| BATF | OCI-Ly3 | GEO | Homo sapiens | GSE56857 | Download (3,428) | ||
| BATF | CD4 | T-cells_d1_anti-BATF | GEO | Homo sapiens | GSE108600 | Download (361) | |
| BATF | CD4 | T-cells_d1_GFP-BATF_anti-GFP | GEO | Homo sapiens | GSE108600 | Download (448) | |
| BATF | CD4 | T-cells_d2_GFP-BATF_anti-BATF | GEO | Homo sapiens | GSE108600 | Download (468) | |
| BATF | CD4 | T-cells_d2_GFP-BATF_anti-GFP | GEO | Homo sapiens | GSE108600 | Download (740) | |
| BATF | GM12878 | ENCODE | Homo sapiens | ENCSR000BGT | Download (80,338) | ||
| BATF | GM12878 | GEO | Homo sapiens | GSE97661 | Download (11,702) | ||
| BATF3 | GM12878 | GEO | Homo sapiens | GSE97661 | Download (9,637) | ||
| BATF3 | KK-1 | BirA | GEO | Homo sapiens | GSE94732 | Download (12,176) | |
| BATF3 | ST-1 | BirA | GEO | Homo sapiens | GSE94732 | Download (6,950) | |
| BBM | Col-0 | Col-0_somatic-embryo | GEO | Arabidopsis thaliana | GSE52400 | Download (16,508) | |
| BBM | Col-0 | Col-0_whole-embryogenic-seedling | GEO | Arabidopsis thaliana | GSE52400 | Download (21,145) | |
| BCL11A | Raji | GEO | Homo sapiens | GSE99019 | Download (426) | ||
| BCL11A | NALM-6 | GEO | Homo sapiens | GSE99019 | Download (563) | ||
| BCL11A | MCF-10A | ENA | Homo sapiens | ERP003925 | Download (7,534) | ||
| BCL11A | MDA-MB-157 | ENA | Homo sapiens | ERP003925 | Download (2,584) | ||
| BCL11A | WA01 | ENCODE | Homo sapiens | ENCSR000BIP | Download (4,096) | ||
| BCL11A | WA01 | ENCODE | Homo sapiens | ENCSR000BMJ | Download (2,697) | ||
| BCL11A | CD34 | Day3_30min | GEO | Homo sapiens | GSE104676 | Download (53,395) | |
| BCL11A | CD34 | Day3_60min | GEO | Homo sapiens | GSE104676 | Download (112,171) | |
| BCL11A | CD34 | Day3_90min | GEO | Homo sapiens | GSE104676 | Download (123,652) | |
| BCL11A | CD34 | Day5_30min | GEO | Homo sapiens | GSE104676 | Download (50,640) | |
| BCL11A | CD34 | Day5_60min | GEO | Homo sapiens | GSE104676 | Download (93,530) | |
| BCL11A | CD34 | Day5_90min | GEO | Homo sapiens | GSE104676 | Download (137,826) | |
| BCL11A | CD34 | Day7_15min | GEO | Homo sapiens | GSE104676 | Download (5,439) | |
| BCL11A | CD34 | Day7_30min | GEO | Homo sapiens | GSE104676 | Download (19,284) | |
| BCL11A | CD34 | Day7_60min | GEO | Homo sapiens | GSE104676 | Download (17,305) | |
| BCL11A | CD34 | Day7_90min | GEO | Homo sapiens | GSE104676 | Download (18,644) | |
| BCL11A | CD34 | Day9_15min | GEO | Homo sapiens | GSE104676 | Download (5,665) | |
| BCL11A | CD34 | Day9_30min | GEO | Homo sapiens | GSE104676 | Download (32,774) | |
| BCL11A | CD34 | Day9_60min | GEO | Homo sapiens | GSE104676 | Download (22,438) | |
| BCL11A | CD34 | Day9_90min | GEO | Homo sapiens | GSE104676 | Download (35,890) | |
| BCL11A | GM12878 | ENCODE | Homo sapiens | ENCSR000BHA | Download (56,037) | ||
| BCL11A | HEK293 | ENCODE | Homo sapiens | ENCSR021DJC | Download (16,144) | ||
| BCL11A | HUDEP-2 | BCL11A-ER-V5 | GEO | Homo sapiens | GSE103445 | Download (13,950) | |
| BCL11A | HUDEP-2 | 30min | GEO | Homo sapiens | GSE104676 | Download (30,605) | |
| BCL11A | HUDEP-2 | 60min | GEO | Homo sapiens | GSE104676 | Download (84,894) | |
| BCL11A | HUDEP-2 | 90min | GEO | Homo sapiens | GSE104676 | Download (52,305) | |
| BCL11A | HUDEP-2 | BCL11A-KO | GEO | Homo sapiens | GSE104676 | Download (109) | |
| BCL11A | HUDEP-2 | CRISPR_115edit_60min | GEO | Homo sapiens | GSE104676 | Download (85,456) | |
| BCL11B | HEK293 | ENCODE | Homo sapiens | ENCSR770PQN | Download (13,696) | ||
| BCL11B | thymus | CD34neg | GEO | Homo sapiens | GSE84677 | Download (39,082) | |
| BCL11B | thymus | CD34pos | GEO | Homo sapiens | GSE84677 | Download (24,020) | |
| BCL3 | GM12878 | ENCODE | Homo sapiens | ENCSR000BNQ | Download (37,840) | ||
| BCL3 | A-549 | ENCODE | Homo sapiens | ENCSR000BQH | Download (38,484) | ||
| BCL6 | Hep-G2 | ENCODE | Homo sapiens | ENCSR759BDG | Download (105,449) | ||
| BCL6 | B-cell | GERMINAL_CENTER | GEO | Homo sapiens | GSE43350 | Download (34,017) | |
| BCL6 | B-cell | GERMINAL_CENTER | GEO | Homo sapiens | GSE68349 | Download (25,046) | |
| BCL6 | OCI-Ly1 | GEO | Homo sapiens | GSE29282 | Download (80,809) | ||
| BCL6 | OCI-Ly7 | GEO | Homo sapiens | GSE44420 | Download (10,136) | ||
| BCL6 | OCI-Ly1 | FA | GEO | Homo sapiens | GSE103125 | Download (254) | |
| BCL6 | OCI-Ly1 | UV | GEO | Homo sapiens | GSE103125 | Download (9,305) | |
| BCL6 | CD4 | GEO | Homo sapiens | GSE59933 | Download (33,830) | ||
| BCL6B | HEK293 | ENCODE | Homo sapiens | ENCSR673SGK | Download (8,475) | ||
| BCLAF1 | GM12878 | ENCODE | Homo sapiens | ENCSR000BJZ | Download (92,667) | ||
| BCLAF1 | GM12878 | ENCODE | Homo sapiens | ENCSR342THD | Download (596) | ||
| BCLAF1 | K-562 | ENCODE | Homo sapiens | ENCSR000BKH | Download (5,856) | ||
| BCLAF1 | K-562 | ENCODE | Homo sapiens | ENCSR492LTS | Download (6,904) | ||
| BCLAF1 | K-562 | ENCODE | Homo sapiens | ENCSR792IYC | Download (4,292) | ||
| BCOR | K-562 | ENCODE | Homo sapiens | ENCSR808AKZ | Download (71,403) | ||
| BCOR | WA01 | GEO | Homo sapiens | GSE104690 | Download (29,734) | ||
| BCOR | WA01 | KDM2B_KO | GEO | Homo sapiens | GSE104690 | Download (12,624) | |
| BCOR | WA01 | RNF2-R | GEO | Homo sapiens | GSE104690 | Download (38,804) | |
| BCOR | WA01 | RNF2-R_Doxycyclin | GEO | Homo sapiens | GSE104690 | Download (42,304) | |
| BCOR | B-cell | GERMINAL_CENTER | GEO | Homo sapiens | GSE43350 | Download (54,417) | |
| BDP1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000DNX | Download (374) | ||
| BDP1 | K-562 | ENCODE | Homo sapiens | ENCSR000DOK | Download (406) | ||
| BDP1 | IMR-90 | GEO | Homo sapiens | GSE47849 | Download (249) | ||
| BEH2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,472) |
| BEH3 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (772) |
| BEH4 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,633) |
| BHLH10 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (860) |
| BHLH104 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (153) |
| BHLH122 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (7,587) |
| BHLH122 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (14,378) |
| BHLH122 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (850) |
| BHLH28 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,236) |
| BHLH74 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (142) |
| BHLHE22 | CAL-1 | GEO | Homo sapiens | GSE43876 | Download (16,404) | ||
| BHLHE40 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZJ | Download (14,383) | ||
| BHLHE40 | GM12878 | ENCODE | Homo sapiens | ENCSR517QHU | Download (45,291) | ||
| BHLHE40 | GM12878 | ENCODE | Homo sapiens | ENCSR987MTA | Download (59,910) | ||
| BHLHE40 | HEK293T | ENCODE | Homo sapiens | ENCSR789GVU | Download (349) | ||
| BHLHE40 | A-549 | ENCODE | Homo sapiens | ENCSR000DYJ | Download (2,863) | ||
| BHLHE40 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BID | Download (3,905) | ||
| BHLHE40 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EDT | Download (33,945) | ||
| BHLHE40 | IMR-90 | ENCODE | Homo sapiens | ENCSR957KYB | Download (63,749) | ||
| BHLHE40 | K-562 | ENCODE | Homo sapiens | ENCSR000EGV | Download (38,904) | ||
| BIM1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (308) |
| BIM1 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (532) |
| BIM2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (5,565) |
| BLC6 | OCI-Ly1 | minusUV_control_ChIP | GEO | Homo sapiens | GSE103125 | Download (100) | |
| BMAL1 | U2OS | GEO | Homo sapiens | GSE69100 | Download (279) | ||
| BMI1 | LNCaP-C4-2 | GEO | Homo sapiens | GSE97831 | Download (144) | ||
| BMI1 | K-562 | ENCODE | Homo sapiens | ENCSR782WRO | Download (10,383) | ||
| BMI1 | MCF-7 | ENCODE | Homo sapiens | ENCSR966YYJ | Download (14,461) | ||
| BMI1 | HEK293T | GEO | Homo sapiens | GSE34774 | Download (55) | ||
| BMI1 | GM12878 | ENCODE | Homo sapiens | ENCSR469WII | Download (12,987) | ||
| BMPR1A | HUVEC-C | GEO | Homo sapiens | GSE60156 | Download (30,454) | ||
| BMY2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,624) |
| BPC1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (297) |
| BPC1 | Col | Col_whole-plant | 30h | GEO | Arabidopsis thaliana | GSE84483 | Download (3,114) |
| BPE | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (142) |
| BPTF | T-47D-MTVL | BPTF | GEO | Homo sapiens | GSE64467 | Download (221) | |
| BRCA1 | U2OS | GEO | Homo sapiens | GSE87324 | Download (3,583) | ||
| BRCA1 | MCF-10A | GEO | Homo sapiens | GSE40591 | Download (16,342) | ||
| BRCA1 | TC-32 | GEO | Homo sapiens | GSE87324 | Download (3,286) | ||
| BRCA1 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZS | Download (486) | ||
| BRCA1 | A-549 | ENCODE | Homo sapiens | ENCSR857KDI | Download (2,325) | ||
| BRCA1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EDY | Download (4,011) | ||
| BRCA1 | K-562 | ENCODE | Homo sapiens | ENCSR223MLH | Download (27,174) | ||
| BRCA1 | WA01 | ENCODE | Homo sapiens | ENCSR000EBX | Download (2,361) | ||
| BRCA1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EDB | Download (13,006) | ||
| BRD1 | HUES-64 | GEO | Homo sapiens | GSE104059 | Download (78,146) | ||
| BRD1 | RKO | GEO | Homo sapiens | GSE47190 | Download (27,145) | ||
| BRD2 | HUVEC-C | GEO | Homo sapiens | GSE60171 | Download (10,803) | ||
| BRD2 | HUVEC-C | MS417 | GEO | Homo sapiens | GSE60171 | Download (332) | |
| BRD2 | NCI-H23 | GEO | Homo sapiens | GSE113714 | Download (18,922) | ||
| BRD2 | SK-MEL-147 | GEO | Homo sapiens | GSE94488 | Download (84,477) | ||
| BRD2 | SK-MEL-147 | JQ1 | GEO | Homo sapiens | GSE94488 | Download (63,783) | |
| BRD2 | NCI-H23 | GEO | Homo sapiens | GSE104481 | Download (16,130) | ||
| BRD2 | HEK293T | GEO | Homo sapiens | GSE39579 | Download (105,921) | ||
| BRD2 | MM1-S | GEO | Homo sapiens | GSE43743 | Download (6,246) | ||
| BRD2 | foreskin | NHM1-5 | GEO | Homo sapiens | GSE94488 | Download (15,575) | |
| BRD3 | MM1-S | GEO | Homo sapiens | GSE43743 | Download (11,884) | ||
| BRD3 | U-87MG | GBM | GEO | Homo sapiens | GSE99171 | Download (20,851) | |
| BRD3 | U-87MG | GBM_dBET6_1d | GEO | Homo sapiens | GSE99171 | Download (7,697) | |
| BRD3 | U-87MG | GBM_dBET6_2h | GEO | Homo sapiens | GSE99171 | Download (8,497) | |
| BRD3 | HEK293T | GEO | Homo sapiens | GSE39579 | Download (33,456) | ||
| BRD3 | A-549 | GEO | Homo sapiens | GSE119863 | Download (9,099) | ||
| BRD3 | HUVEC-C | GEO | Homo sapiens | GSE60171 | Download (5,321) | ||
| BRD3 | HUVEC-C | MS417 | GEO | Homo sapiens | GSE60171 | Download (1,729) | |
| BRD4 | hESC | GEO | Homo sapiens | GSE33281 | Download (63,822) | ||
| BRD4 | HFOB | DIFF | GEO | Homo sapiens | GSE82295 | Download (18,667) | |
| BRD4 | HT29 | GEO | Homo sapiens | GSE73319 | Download (28,261) | ||
| BRD4 | Jurkat | GEO | Homo sapiens | GSE83777 | Download (47,877) | ||
| BRD4 | KOPTK1 | GEO | Homo sapiens | GSE54379 | Download (39,100) | ||
| BRD4 | KOPTK1 | E | GEO | Homo sapiens | GSE54379 | Download (37,789) | |
| BRD4 | LP1 | GEO | Homo sapiens | GSE71909 | Download (1,247) | ||
| BRD4 | LP1 | CPI203 | GEO | Homo sapiens | GSE71909 | Download (181) | |
| BRD4 | LP1 | SGCCBP300 | GEO | Homo sapiens | GSE71909 | Download (651) | |
| BRD4 | MPNST | DMSO | GEO | Homo sapiens | GSE62499 | Download (95,431) | |
| BRD4 | MPNST | PDJQ | GEO | Homo sapiens | GSE62499 | Download (45,258) | |
| BRD4 | SEM | GEO | Homo sapiens | GSE83671 | Download (34,432) | ||
| BRD4 | SGBS | GEO | Homo sapiens | GSE64233 | Download (6,101) | ||
| BRD4 | SGBS | TNF | GEO | Homo sapiens | GSE64233 | Download (8,194) | |
| BRD4 | SUM1315 | DMSO | GEO | Homo sapiens | GSE63581 | Download (26,930) | |
| BRD4 | SUM149 | DMSO | GEO | Homo sapiens | GSE63581 | Download (14,110) | |
| BRD4 | SUM159 | GEO | Homo sapiens | GSE63581 | Download (623) | ||
| BRD4 | SUM159 | RES | GEO | Homo sapiens | GSE63581 | Download (1,935) | |
| BRD4 | SUM185 | DMSO | GEO | Homo sapiens | GSE63581 | Download (16,399) | |
| BRD4 | SW480 | GEO | Homo sapiens | GSE110473 | Download (39,008) | ||
| BRD4 | SW480 | 16h_TNF-a | GEO | Homo sapiens | GSE110473 | Download (34,110) | |
| BRD4 | SW480 | GEO | Homo sapiens | GSE73319 | Download (25,012) | ||
| BRD4 | HCC1806 | 100nMtrametinib_24h | GEO | Homo sapiens | GSE87418 | Download (30,891) | |
| BRD4 | HCC1806 | 100nMtrametinib300nMJQ1_24h | GEO | Homo sapiens | GSE87418 | Download (19,741) | |
| BRD4 | HCC1806 | 300nMJQ1_24h | GEO | Homo sapiens | GSE87418 | Download (12,839) | |
| BRD4 | HCC1806 | DMSO_24h | GEO | Homo sapiens | GSE87418 | Download (17,101) | |
| BRD4 | HUVEC-C | GEO | Homo sapiens | GSE53998 | Download (21,185) | ||
| BRD4 | HUVEC-C | TNF | GEO | Homo sapiens | GSE53998 | Download (22,776) | |
| BRD4 | HUVEC-C | TNF_JQ1 | GEO | Homo sapiens | GSE53998 | Download (6,308) | |
| BRD4 | HUVEC-C | GEO | Homo sapiens | GSE60171 | Download (4,085) | ||
| BRD4 | HUVEC-C | MS417 | GEO | Homo sapiens | GSE60171 | Download (84) | |
| BRD4 | HUVEC-C | modETS1 | GEO | Homo sapiens | GSE93030 | Download (16,570) | |
| BRD4 | HUVEC-C | modGFP | GEO | Homo sapiens | GSE93030 | Download (13,609) | |
| BRD4 | IMR-90 | PROLIF | GEO | Homo sapiens | GSE74238 | Download (621) | |
| BRD4 | IMR-90 | QUIES | GEO | Homo sapiens | GSE74238 | Download (73,760) | |
| BRD4 | IMR-90 | SENES | GEO | Homo sapiens | GSE74238 | Download (27,037) | |
| BRD4 | MCF-10A | GEO | Homo sapiens | GSE72931 | Download (82,441) | ||
| BRD4 | MCF-7 | GEO | Homo sapiens | GSE55921 | Download (10,180) | ||
| BRD4 | MCF-7 | E2 | GEO | Homo sapiens | GSE55921 | Download (9,678) | |
| BRD4 | MDA-MB-436 | DMSO | GEO | Homo sapiens | GSE63581 | Download (6,192) | |
| BRD4 | MOLM-14 | CA25 | GEO | Homo sapiens | GSE65138 | Download (26,007) | |
| BRD4 | MOLM-14 | DMSO | GEO | Homo sapiens | GSE65138 | Download (41,965) | |
| BRD4 | MOLM-14 | IBET | GEO | Homo sapiens | GSE65138 | Download (40,682) | |
| BRD4 | MOLT-4 | DMSO | GEO | Homo sapiens | GSE79288 | Download (17,089) | |
| BRD4 | Mutu-1 | JQ1 | GEO | Homo sapiens | GSE84213 | Download (9,066) | |
| BRD4 | Mutu-1 | vehicle | GEO | Homo sapiens | GSE84213 | Download (30,292) | |
| BRD4 | MV4-11 | DMSO | GEO | Homo sapiens | GSE71776 | Download (38,449) | |
| BRD4 | MV4-11 | IBET | GEO | Homo sapiens | GSE71776 | Download (38,512) | |
| BRD4 | MV4-11 | IBET_SGC | GEO | Homo sapiens | GSE71776 | Download (27,606) | |
| BRD4 | MV4-11 | SGC | GEO | Homo sapiens | GSE71776 | Download (61,906) | |
| BRD4 | NCI-H2171 | GEO | Homo sapiens | GSE42355 | Download (91,761) | ||
| BRD4 | NCI-H2171 | DMSO | GEO | Homo sapiens | GSE49224 | Download (6,186) | |
| BRD4 | NCI-H2171 | JQ1 | GEO | Homo sapiens | GSE49224 | Download (42) | |
| BRD4 | OCI-Ly1 | DMSO | GEO | Homo sapiens | GSE53601 | Download (5,846) | |
| BRD4 | OCI-Ly1 | JQ1 | GEO | Homo sapiens | GSE53601 | Download (13,561) | |
| BRD4 | OVCAR-3 | DMSO | GEO | Homo sapiens | GSE77568 | Download (14,203) | |
| BRD4 | OVCAR-3 | JQ1 | GEO | Homo sapiens | GSE77568 | Download (375) | |
| BRD4 | P493-6 | MYC_0H | GEO | Homo sapiens | GSE42262 | Download (4,023) | |
| BRD4 | P493-6 | MYC_1H | GEO | Homo sapiens | GSE42262 | Download (8,260) | |
| BRD4 | P493-6 | MYC_24H | GEO | Homo sapiens | GSE42262 | Download (2,003) | |
| BRD4 | RH4 | GEO | Homo sapiens | GSE83726 | Download (16,567) | ||
| BRD4 | SUM159PT | 100nMtrametinib_1h | GEO | Homo sapiens | GSE87418 | Download (61,944) | |
| BRD4 | SUM159PT | 100nMtrametinib1uMSGCCBP30_24h | GEO | Homo sapiens | GSE87418 | Download (87,444) | |
| BRD4 | SUM159PT | 100nMtrametinib_24h | GEO | Homo sapiens | GSE87418 | Download (85,237) | |
| BRD4 | SUM159PT | 100nMtrametinib300nMJIB04_24h | GEO | Homo sapiens | GSE87418 | Download (96,580) | |
| BRD4 | SUM159PT | 100nMtrametinib300nMJQ1_24h | GEO | Homo sapiens | GSE87418 | Download (37,418) | |
| BRD4 | SUM159PT | 100nMtrametinib30nMbortezomib_8h | GEO | Homo sapiens | GSE87418 | Download (83,542) | |
| BRD4 | SUM159PT | 100nMtrametinib_48h | GEO | Homo sapiens | GSE87418 | Download (84,845) | |
| BRD4 | SUM159PT | 100nMtrametinib_4h | GEO | Homo sapiens | GSE87418 | Download (56,375) | |
| BRD4 | SUM159PT | 100nMtrametinib_72h | GEO | Homo sapiens | GSE87418 | Download (78,003) | |
| BRD4 | SUM159PT | 100nMtrametinib_8h | GEO | Homo sapiens | GSE87418 | Download (64,081) | |
| BRD4 | SUM159PT | 300nMJQ1_24h | GEO | Homo sapiens | GSE87418 | Download (23,791) | |
| BRD4 | SUM159PT | 30nMbortezomib_8h | GEO | Homo sapiens | GSE87418 | Download (81,674) | |
| BRD4 | SUM159PT | DMSO_24h | GEO | Homo sapiens | GSE87418 | Download (58,621) | |
| BRD4 | SUM159PT | DMSO_48h | GEO | Homo sapiens | GSE87418 | Download (52,429) | |
| BRD4 | SUM159PT | DMSO_72h | GEO | Homo sapiens | GSE87418 | Download (38,067) | |
| BRD4 | SUM159PT | DMSO_8h | GEO | Homo sapiens | GSE87418 | Download (59,138) | |
| BRD4 | SUM159PT | Dox_48h | GEO | Homo sapiens | GSE87418 | Download (55,813) | |
| BRD4 | SUM159PT | shMYC_BRD4_100nMtrametinib_48h | GEO | Homo sapiens | GSE87418 | Download (93,297) | |
| BRD4 | SUM159PT | shMYC_DMSO_48h | GEO | Homo sapiens | GSE87418 | Download (58,667) | |
| BRD4 | SUM159PT | shMYC_dox_48h | GEO | Homo sapiens | GSE87418 | Download (72,660) | |
| BRD4 | SUM229PE | neg_30nMtrametinib_24h | GEO | Homo sapiens | GSE87418 | Download (34,743) | |
| BRD4 | SUM229PE | neg_DMSO_24h | GEO | Homo sapiens | GSE87418 | Download (27,764) | |
| BRD4 | SUM229PE | pos_30nMtrametinib_24h | GEO | Homo sapiens | GSE87418 | Download (21,232) | |
| BRD4 | SUM229PE | pos_DMSO_24h | GEO | Homo sapiens | GSE87418 | Download (29,759) | |
| BRD4 | T-47D | DMSO | GEO | Homo sapiens | GSE63581 | Download (26,136) | |
| BRD4 | K-562 | ENCODE | Homo sapiens | ENCSR583ACG | Download (9,288) | ||
| BRD4 | MCF-10A | ENA | Homo sapiens | ERP003925 | Download (38,485) | ||
| BRD4 | MDA-MB-157 | ENA | Homo sapiens | ERP003925 | Download (21,240) | ||
| BRD4 | MDA-MB-231 | ENA | Homo sapiens | ERP003925 | Download (7,002) | ||
| BRD4 | NCI-H2171 | GEO | Homo sapiens | GSE101821 | Download (5,639) | ||
| BRD4 | T-47D | ENA | Homo sapiens | ERP003925 | Download (21,603) | ||
| BRD4 | MV4-11-B | BI00894999_100nM_4h | GEO | Homo sapiens | GSE101821 | Download (64,851) | |
| BRD4 | MV4-11-B | BI00894999_1nM_4h | GEO | Homo sapiens | GSE101821 | Download (82,832) | |
| BRD4 | MV4-11-B | BI00894999_35nM_4h | GEO | Homo sapiens | GSE101821 | Download (68,421) | |
| BRD4 | MV4-11-B | DMSO | GEO | Homo sapiens | GSE101821 | Download (49,432) | |
| BRD4 | LNAR | Enz | GEO | Homo sapiens | GSE103449 | Download (493) | |
| BRD4 | LREX | GEO | Homo sapiens | GSE103449 | Download (168) | ||
| BRD4 | OCI-AML3 | GEO | Homo sapiens | GSE104745 | Download (16,475) | ||
| BRD4 | OCI-AML3 | JQ1_500nM_24h | GEO | Homo sapiens | GSE104745 | Download (1,557) | |
| BRD4 | BE2C | GEO | Homo sapiens | GSE80151 | Download (20,935) | ||
| BRD4 | CD4 | GEO | Homo sapiens | GSE33281 | Download (88,615) | ||
| BRD4 | CD4 | JG1 | GEO | Homo sapiens | GSE33281 | Download (72,298) | |
| BRD4 | CD4 | TH1_BAY | GEO | Homo sapiens | GSE62482 | Download (19,341) | |
| BRD4 | CD4 | TH1_DMSO | GEO | Homo sapiens | GSE62482 | Download (35,557) | |
| BRD4 | DND41 | GEO | Homo sapiens | GSE54379 | Download (10,961) | ||
| BRD4 | DND41 | E | GEO | Homo sapiens | GSE54379 | Download (23,953) | |
| BRD4 | HCC1395 | GEO | Homo sapiens | GSE63581 | Download (65,899) | ||
| BRD4 | HCC1395 | JQ1 | GEO | Homo sapiens | GSE63581 | Download (30,139) | |
| BRD4 | HCT15 | GEO | Homo sapiens | GSE73319 | Download (33,224) | ||
| BRD4 | HEK293T | GEO | Homo sapiens | GSE39579 | Download (14,073) | ||
| BRD4 | HEK293T | GEO | Homo sapiens | GSE50491 | Download (46) | ||
| BRD4 | HEK293T | GEO | Homo sapiens | GSE51633 | Download (26,688) | ||
| BRD4 | HeLa | GEO | Homo sapiens | GSE51633 | Download (17,143) | ||
| BRD4 | CLL | patient1 | GEO | Homo sapiens | GSE109411 | Download (37,225) | |
| BRD4 | CLL | patient1_4h_CpG | GEO | Homo sapiens | GSE109411 | Download (35,502) | |
| BRD4 | CLL | patient1_4h_CpG_BET | GEO | Homo sapiens | GSE109411 | Download (19,348) | |
| BRD4 | CLL | patient2 | GEO | Homo sapiens | GSE109411 | Download (55,888) | |
| BRD4 | CLL | patient2_4h_CpG | GEO | Homo sapiens | GSE109411 | Download (30,078) | |
| BRD4 | CLL | patient2_4h_CpG_BET | GEO | Homo sapiens | GSE109411 | Download (36,089) | |
| BRD4 | CLL | patient3 | GEO | Homo sapiens | GSE109411 | Download (39,129) | |
| BRD4 | CLL | patient3_4h_CpG | GEO | Homo sapiens | GSE109411 | Download (47,912) | |
| BRD4 | CLL | patient3_4h_CpG_BET | GEO | Homo sapiens | GSE109411 | Download (24,527) | |
| BRD4 | CLL | patient4 | GEO | Homo sapiens | GSE109411 | Download (38,394) | |
| BRD4 | CLL | patient4_4h_CpG | GEO | Homo sapiens | GSE109411 | Download (33,418) | |
| BRD4 | CLL | patient4_4h_CpG_BET | GEO | Homo sapiens | GSE109411 | Download (21,630) | |
| BRD4 | MM1-S | DMSO | GEO | Homo sapiens | GSE42161 | Download (26,518) | |
| BRD4 | MM1-S | JQ1 | GEO | Homo sapiens | GSE42161 | Download (690) | |
| BRD4 | MM1-S | JQ1_500NM | GEO | Homo sapiens | GSE42355 | Download (41,681) | |
| BRD4 | MM1-S | JQ1_50NM | GEO | Homo sapiens | GSE42355 | Download (50,254) | |
| BRD4 | MM1-S | JQ1_5NM | GEO | Homo sapiens | GSE42355 | Download (57,587) | |
| BRD4 | MM1-S | JQ1_5UM | GEO | Homo sapiens | GSE42355 | Download (36,962) | |
| BRD4 | U-87MG | GEO | Homo sapiens | GSE42355 | Download (83,853) | ||
| BRD4 | MM1-S | GEO | Homo sapiens | GSE43743 | Download (46,579) | ||
| BRD4 | MM1-S | GEO | Homo sapiens | GSE45984 | Download (26,523) | ||
| BRD4 | MM1-S | JQ1_150NM | GEO | Homo sapiens | GSE49224 | Download (42,778) | |
| BRD4 | U-87MG | DMSO | GEO | Homo sapiens | GSE49224 | Download (106,168) | |
| BRD4 | U-87MG | JQ1 | GEO | Homo sapiens | GSE49224 | Download (109,501) | |
| BRD4 | HCT-116 | GEO | Homo sapiens | GSE57628 | Download (12,886) | ||
| BRD4 | HCT-116 | JQ1 | GEO | Homo sapiens | GSE57628 | Download (3,018) | |
| BRD4 | COLO-205 | GEO | Homo sapiens | GSE73319 | Download (30,284) | ||
| BRD4 | COLO-320 | GEO | Homo sapiens | GSE73319 | Download (9,327) | ||
| BRD4 | COLO-741 | GEO | Homo sapiens | GSE73319 | Download (57,154) | ||
| BRD4 | HCT-116 | GEO | Homo sapiens | GSE73319 | Download (7,073) | ||
| BRD4 | GEN2-2 | GEO | Homo sapiens | GSE76147 | Download (340) | ||
| BRD4 | SK-N-BE2-C | GEO | Homo sapiens | GSE80151 | Download (21,438) | ||
| BRD4 | Hs-352-Sk | GEO | Homo sapiens | GSE83725 | Download (4,326) | ||
| BRD4 | Hs-352-Sk | PAX3-FOXO1-vector | GEO | Homo sapiens | GSE83725 | Download (14,616) | |
| BRD4 | retina | AB1-FW13 | GEO | Homo sapiens | GSE86981 | Download (12,888) | |
| BRD4 | retina | AB1-FW15 | GEO | Homo sapiens | GSE86981 | Download (83,875) | |
| BRD4 | retina | AB1-FW20 | GEO | Homo sapiens | GSE86981 | Download (31,402) | |
| BRD4 | retina | AB1-FW23 | GEO | Homo sapiens | GSE86981 | Download (33,473) | |
| BRD4 | retina | AB1-RB | GEO | Homo sapiens | GSE86981 | Download (74,266) | |
| BRD4 | LNCaP-C4-2 | EV | GEO | Homo sapiens | GSE88871 | Download (5,943) | |
| BRD4 | LNCaP-C4-2 | EV_JQ1 | GEO | Homo sapiens | GSE88871 | Download (342) | |
| BRD4 | LNCaP-C4-2 | F133V | GEO | Homo sapiens | GSE88871 | Download (80,615) | |
| BRD4 | LNCaP-C4-2 | F133V_JQ1 | GEO | Homo sapiens | GSE88871 | Download (45,419) | |
| BRD4 | LNCaP-C4-2 | OE | GEO | Homo sapiens | GSE88871 | Download (72,010) | |
| BRD4 | CHL-1 | GEO | Homo sapiens | GSE95585 | Download (34,992) | ||
| BRD4 | CHL-1 | BAY123897 | GEO | Homo sapiens | GSE95585 | Download (7,592) | |
| BRD4 | CHL-1 | OTX015 | GEO | Homo sapiens | GSE95585 | Download (5,855) | |
| BRD4 | GM15850 | DMSO | GEO | Homo sapiens | GSE99402 | Download (27,908) | |
| BRD4 | GM15850 | PA1_JQ1 | GEO | Homo sapiens | GSE99402 | Download (17,908) | |
| BRD4 | GM15850 | Syn-TEF1 | GEO | Homo sapiens | GSE99402 | Download (30,667) | |
| BRD4 | K-562 | DMSO | GEO | Homo sapiens | GSE99178 | Download (34,572) | |
| BRD4 | K-562 | JQ1_2h | GEO | Homo sapiens | GSE99178 | Download (21,844) | |
| BRD4 | K-562 | JQ1_6h | GEO | Homo sapiens | GSE99178 | Download (15,045) | |
| BRD4 | KK-1 | DMSO | GEO | Homo sapiens | GSE94732 | Download (5,164) | |
| BRD4 | KK-1 | JQ1 | GEO | Homo sapiens | GSE94732 | Download (3,288) | |
| BRD4 | NMC24335 | GEO | Homo sapiens | GSE96775 | Download (25,601) | ||
| BRD4 | BT-474 | ENA | Homo sapiens | ERP010664 | Download (6,490) | ||
| BRD4 | BT-474 | INHHDAC | ENA | Homo sapiens | ERP010664 | Download (3,809) | |
| BRD4 | BCBL-1 | TREx-F3H3-K-Rt | GEO | Homo sapiens | GSE103395 | Download (592) | |
| BRD4 | Hep-G2 | ENCODE | Homo sapiens | ENCSR514EOE | Download (7,292) | ||
| BRD9 | K-562 | ENCODE | Homo sapiens | ENCSR177XCS | Download (20,602) | ||
| BRF1 | K-562 | ENCODE | Homo sapiens | ENCSR000DOJ | Download (158) | ||
| BRF1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000DNW | Download (115) | ||
| BRF2 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000DNV | Download (13) | ||
| BRF2 | K-562 | ENCODE | Homo sapiens | ENCSR000DOC | Download (23) | ||
| BRF2 | IMR-90 | TERT | GEO | Homo sapiens | GSE38303 | Download (2,800) | |
| BRG1 | LNCaP | r1881 | GEO | Homo sapiens | GSE94682 | Download (11,962) | |
| BRG1 | WA09 | heat-shock | GEO | Homo sapiens | GSE105028 | Download (3,058) | |
| BRG1 | HS-SY-2 | HA-tagged | GEO | Homo sapiens | GSE108926 | Download (57,289) | |
| BZIP28 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (817) |
| BZIP68 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (4,187) |
| BZR1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (206) |
| C11orf30 | K-562 | ENCODE | Homo sapiens | ENCSR350XWY | Download (83,655) | ||
| C17orf49 | HeLa | GEO | Homo sapiens | GSE20303 | Download (208) | ||
| CARM1 | H3396 | E2 | GEO | Homo sapiens | GSE32349 | Download (19) | |
| CBFA2T2 | K-562 | ENCODE | Homo sapiens | ENCSR699PVC | Download (26,659) | ||
| CBFA2T2 | NCCIT | GEO | Homo sapiens | GSE71675 | Download (38,716) | ||
| CBFA2T3 | K-562 | ENCODE | Homo sapiens | ENCSR697YLJ | Download (55,031) | ||
| CBFB | ME-1 | GEO | Homo sapiens | GSE46044 | Download (54,240) | ||
| CBFB | GM12878 | ENCODE | Homo sapiens | ENCSR860UHK | Download (33,173) | ||
| CBFB | SKNO1 | GEO | Homo sapiens | GSE23730 | Download (17,247) | ||
| CBFB | SaOS-2 | GEO | Homo sapiens | GSE76937 | Download (11,845) | ||
| CBX1 | K-562 | ENCODE | Homo sapiens | ENCSR948QLZ | Download (33,193) | ||
| CBX1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR819WZE | Download (69,306) | ||
| CBX2 | K-562 | ENCODE | Homo sapiens | ENCSR000ATU | Download (33,869) | ||
| CBX2 | HEK293T | GEO | Homo sapiens | GSE34774 | Download (14,103) | ||
| CBX3 | HCT-116 | ENCODE | Homo sapiens | ENCSR000BUH | Download (27,745) | ||
| CBX3 | T-47D-MTVL | GEO | Homo sapiens | GSE64467 | Download (1,307) | ||
| CBX3 | T-47D-MTVL | BPTF | GEO | Homo sapiens | GSE64467 | Download (370) | |
| CBX3 | K-562 | ENCODE | Homo sapiens | ENCSR000ATV | Download (296) | ||
| CBX3 | K-562 | ENCODE | Homo sapiens | ENCSR000BRT | Download (93,758) | ||
| CBX4 | HEK293T | GEO | Homo sapiens | GSE53495 | Download (921) | ||
| CBX5 | GM12878 | ENCODE | Homo sapiens | ENCSR372GIN | Download (12,750) | ||
| CBX5 | K-562 | ENCODE | Homo sapiens | ENCSR272JAT | Download (9,504) | ||
| CBX8 | A-549 | ENCODE | Homo sapiens | ENCSR616MOB | Download (11,470) | ||
| CBX8 | K-562 | ENCODE | Homo sapiens | ENCSR000ATW | Download (56,663) | ||
| CC2D1A | K-562 | ENCODE | Homo sapiens | ENCSR343IFJ | Download (20,325) | ||
| CCA1 | Col-0 | Col-0_seedling | 14d | GEO | Arabidopsis thaliana | GSE67903 | Download (5,046) |
| CCA1 | Ws-2 | Ws-2_seedling | 14d-LD | GEO | Arabidopsis thaliana | GSE70533 | Download (5,527) |
| CCA1 | Ws-2 | Ws-2_seedling | 14d-LL | GEO | Arabidopsis thaliana | GSE70533 | Download (2,699) |
| CCA1 | Ws-2 | Ws-2_seedling | 14d-LL-ZT14 | GEO | Arabidopsis thaliana | GSE70533 | Download (418) |
| CCAR2 | K-562 | ENCODE | Homo sapiens | ENCSR598GER | Download (1,089) | ||
| CCAR2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR247XFV | Download (28,028) | ||
| CCNT2 | K-562 | ENCODE | Homo sapiens | ENCSR000DOA | Download (22,622) | ||
| CD74 | CLL | p1 | GEO | Homo sapiens | GSE88955 | Download (5,640) | |
| CD74 | CLL | p2 | GEO | Homo sapiens | GSE88955 | Download (1,794) | |
| CD74 | CLL | p3 | GEO | Homo sapiens | GSE88955 | Download (1,921) | |
| CD74 | CLL | p4 | GEO | Homo sapiens | GSE88955 | Download (8,978) | |
| CDC5L | K-562 | ENCODE | Homo sapiens | ENCSR121PFY | Download (4,936) | ||
| CDF3 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,591) |
| CDF5 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,433) |
| CDK2 | T-47D | GEO | Homo sapiens | GSE53855 | Download (276) | ||
| CDK2 | T-47D | PROGESTIN_30M | GEO | Homo sapiens | GSE53855 | Download (927) | |
| CDK2 | T-47D | PROGESTIN_5M | GEO | Homo sapiens | GSE53855 | Download (2,176) | |
| CDK6 | KB | GEO | Homo sapiens | GSE52469 | Download (10,672) | ||
| CDK6 | KB | IL | GEO | Homo sapiens | GSE52469 | Download (61,467) | |
| CDK7 | Jurkat | GEO | Homo sapiens | GSE50622 | Download (15,933) | ||
| CDK7 | Jurkat | DMSO | GEO | Homo sapiens | GSE60027 | Download (23,501) | |
| CDK7 | Jurkat | THZ2102 | GEO | Homo sapiens | GSE60027 | Download (11,002) | |
| CDK7 | Jurkat | GEO | Homo sapiens | GSE83777 | Download (32,533) | ||
| CDK7 | SK-MEL-147 | GEO | Homo sapiens | GSE45984 | Download (8,955) | ||
| CDK7 | MM1-S | GEO | Homo sapiens | GSE45984 | Download (1,183) | ||
| CDK8 | HCT-116 | HYPO | GEO | Homo sapiens | GSE38258 | Download (101,990) | |
| CDK8 | HCT-116 | NOMO | GEO | Homo sapiens | GSE38258 | Download (23,307) | |
| CDK8 | MM1-S | GEO | Homo sapiens | GSE43743 | Download (3,808) | ||
| CDK8 | SW480 | GEO | Homo sapiens | GSE53602 | Download (12,729) | ||
| CDK8 | K-562 | GEO | Homo sapiens | GSE65138 | Download (2,114) | ||
| CDK8 | MOLM-14 | GEO | Homo sapiens | GSE65138 | Download (25,519) | ||
| CDK8 | MOLM-14 | DMSO | GEO | Homo sapiens | GSE65138 | Download (124,396) | |
| CDK8 | MOLM-14 | IBET | GEO | Homo sapiens | GSE65138 | Download (59,653) | |
| CDK8 | MV4-11 | GEO | Homo sapiens | GSE65138 | Download (5,693) | ||
| CDK8 | SET-2 | GEO | Homo sapiens | GSE65138 | Download (27,735) | ||
| CDK9 | A-375 | A771726 | GEO | Homo sapiens | GSE57431 | Download (4,762) | |
| CDK9 | A-375 | DMSO | GEO | Homo sapiens | GSE57431 | Download (10,041) | |
| CDK9 | A-375 | A771726 | GEO | Homo sapiens | GSE68052 | Download (4,170) | |
| CDK9 | A-375 | DMSO | GEO | Homo sapiens | GSE68052 | Download (7,104) | |
| CDK9 | MOLT-4 | DMSO | GEO | Homo sapiens | GSE79288 | Download (21,975) | |
| CDK9 | MOLT-4 | JQ1 | GEO | Homo sapiens | GSE79288 | Download (19,263) | |
| CDK9 | MV4-11 | DMSO | GEO | Homo sapiens | GSE82116 | Download (36,719) | |
| CDK9 | P493-6 | GEO | Homo sapiens | GSE36354 | Download (7,633) | ||
| CDK9 | P493-6 | CMYC_1H | GEO | Homo sapiens | GSE36354 | Download (1,444) | |
| CDK9 | P493-6 | CMYC_24H | GEO | Homo sapiens | GSE36354 | Download (255) | |
| CDK9 | MM1-S | DMSO | GEO | Homo sapiens | GSE42161 | Download (35,623) | |
| CDK9 | MM1-S | JQ1 | GEO | Homo sapiens | GSE42161 | Download (116) | |
| CDK9 | MM1-S | DMSO | GEO | Homo sapiens | GSE42355 | Download (6,940) | |
| CDK9 | MM1-S | JQ1_500NM | GEO | Homo sapiens | GSE42355 | Download (42,847) | |
| CDK9 | MM1-S | JQ1_5000NM | GEO | Homo sapiens | GSE49224 | Download (34,873) | |
| CDK9 | MM1-S | JQ1_50NM | GEO | Homo sapiens | GSE49224 | Download (28,650) | |
| CDK9 | HCT-116 | SHCTR | GEO | Homo sapiens | GSE70408 | Download (120) | |
| CDK9 | HCT-116 | GEO | Homo sapiens | GSE72622 | Download (29,629) | ||
| CDK9 | BT-474 | ENA | Homo sapiens | ERP010664 | Download (5,615) | ||
| CDK9 | BT-474 | INHHDAC | ENA | Homo sapiens | ERP010664 | Download (5,643) | |
| CDK9 | CD4 | TH1_BAY | GEO | Homo sapiens | GSE62482 | Download (127) | |
| CDK9 | CD4 | TH1_DMSO | GEO | Homo sapiens | GSE62482 | Download (158) | |
| CDK9 | HEK293T | SIBRD4 | GEO | Homo sapiens | GSE51633 | Download (7,697) | |
| CDK9 | HEK293T | SICTR | GEO | Homo sapiens | GSE51633 | Download (15,608) | |
| CDK9 | HEK293T | SIJMJD6 | GEO | Homo sapiens | GSE51633 | Download (2,556) | |
| CDK9-HEXIM1 | A-375 | GEO | Homo sapiens | GSE68047 | Download (2,446) | ||
| CDX2 | COLO-320 | GEO | Homo sapiens | GSE30026 | Download (18,518) | ||
| CDX2 | CACO2 | DIFF | GEO | Homo sapiens | GSE23436 | Download (29,853) | |
| CDX2 | CACO2 | PROLIF | GEO | Homo sapiens | GSE23436 | Download (4,725) | |
| CDX2 | LS180 | GEO | Homo sapiens | GSE31939 | Download (37,684) | ||
| CDX2 | LS180 | 125 | GEO | Homo sapiens | GSE31939 | Download (47,037) | |
| CEBPA | SGBS | GEO | Homo sapiens | GSE41629 | Download (8,800) | ||
| CEBPA | Kasumi-1 | SICTR | GEO | Homo sapiens | GSE60130 | Download (1,848) | |
| CEBPA | Kasumi-1 | SIRUNX1ETO | GEO | Homo sapiens | GSE60130 | Download (14,853) | |
| CEBPA | MV4-11 | GEO | Homo sapiens | GSE88746 | Download (64,492) | ||
| CEBPA | SKH1 | 10d | GEO | Homo sapiens | GSE87283 | Download (19,415) | |
| CEBPA | SKH1 | RUNX1-EVI1_KD | GEO | Homo sapiens | GSE87283 | Download (31,803) | |
| CEBPA | Kasumi-1 | GEO | Homo sapiens | GSE102697 | Download (8,903) | ||
| CEBPA | Kasumi-1 | CEBPA-ER | GEO | Homo sapiens | GSE102697 | Download (79,510) | |
| CEBPA | Kasumi-1 | CEBPA-ER_E2 | GEO | Homo sapiens | GSE102697 | Download (67,917) | |
| CEBPA | Kasumi-1 | E2 | GEO | Homo sapiens | GSE102697 | Download (11,376) | |
| CEBPA | SKH1 | GEO | Homo sapiens | GSE102697 | Download (6,382) | ||
| CEBPA | SKH1 | CEBPA-ER | GEO | Homo sapiens | GSE102697 | Download (22,776) | |
| CEBPA | SKH1 | CEBPA-ER_E2 | GEO | Homo sapiens | GSE102697 | Download (21,099) | |
| CEBPA | SKH1 | E2 | GEO | Homo sapiens | GSE102697 | Download (11,601) | |
| CEBPA | U-937 | ENA | Homo sapiens | ERP008568 | Download (29,126) | ||
| CEBPA | liver | ENA | Homo sapiens | ERP002306 | Download (37,947) | ||
| CEBPA | Hep-G2 | ENCODE | Homo sapiens | ENCSR142IGM | Download (138,802) | ||
| CEBPA | Hep-G2 | ENA | Homo sapiens | ERP000209 | Download (41,468) | ||
| CEBPB | GM12878 | ENCODE | Homo sapiens | ENCSR000BRX | Download (3,593) | ||
| CEBPB | GM12878 | ENCODE | Homo sapiens | ENCSR681NOM | Download (3,639) | ||
| CEBPB | Ishikawa | ENCODE | Homo sapiens | ENCSR000BTT | Download (29,501) | ||
| CEBPB | A-549 | ENCODE | Homo sapiens | ENCSR000BUB | Download (51,051) | ||
| CEBPB | A-549 | ENCODE | Homo sapiens | ENCSR000DYI | Download (98,907) | ||
| CEBPB | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BQI | Download (37,368) | ||
| CEBPB | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEE | Download (140,679) | ||
| CEBPB | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEX | Download (35,293) | ||
| CEBPB | HL-60 | CEBPB_overexpressed | GEO | Homo sapiens | GSE100486 | Download (32,711) | |
| CEBPB | HL-60 | GEO | Homo sapiens | GSE107553 | Download (8,939) | ||
| CEBPB | IMR-90 | ENCODE | Homo sapiens | ENCSR000EFM | Download (193,528) | ||
| CEBPB | K-562 | ENCODE | Homo sapiens | ENCSR000BRQ | Download (52,230) | ||
| CEBPB | K-562 | ENCODE | Homo sapiens | ENCSR000EHE | Download (73,647) | ||
| CEBPB | K-562 | ENCODE | Homo sapiens | ENCSR416QLJ | Download (91,984) | ||
| CEBPB | HFOB | DIFF | GEO | Homo sapiens | GSE82295 | Download (74,204) | |
| CEBPB | hMSC | GEO | Homo sapiens | GSE68864 | Download (30,587) | ||
| CEBPB | hMSC | DMI_HI | GEO | Homo sapiens | GSE68864 | Download (82,038) | |
| CEBPB | hMSC | DMI_LOW | GEO | Homo sapiens | GSE68864 | Download (86,583) | |
| CEBPB | LS180 | GEO | Homo sapiens | GSE31939 | Download (116,021) | ||
| CEBPB | LS180 | 125 | GEO | Homo sapiens | GSE31939 | Download (142,595) | |
| CEBPB | monocyte | GEO | Homo sapiens | GSE31621 | Download (3,452) | ||
| CEBPB | monocyte | MACROPHAGE | GEO | Homo sapiens | GSE31621 | Download (17,488) | |
| CEBPB | MV4-11 | GEO | Homo sapiens | GSE88746 | Download (73,015) | ||
| CEBPB | SUM159PT | 100nMtrametinib_24h | GEO | Homo sapiens | GSE87418 | Download (90,333) | |
| CEBPB | SUM159PT | 100nMtrametinib300nMJQ1_24h | GEO | Homo sapiens | GSE87418 | Download (119,711) | |
| CEBPB | SUM159PT | 300nMJQ1_24h | GEO | Homo sapiens | GSE87418 | Download (101,107) | |
| CEBPB | SUM159PT | DMSO_24h | GEO | Homo sapiens | GSE87418 | Download (84,201) | |
| CEBPB | monocyte | GEO | Homo sapiens | GSE98367 | Download (26,244) | ||
| CEBPB | monocyte | INFg | GEO | Homo sapiens | GSE98367 | Download (40,960) | |
| CEBPB | MCF-7 | ENCODE | Homo sapiens | ENCSR000BSR | Download (68,269) | ||
| CEBPB | THP-1 | eGFP-Pam3csk-0h | GEO | Homo sapiens | GSE103477 | Download (17,660) | |
| CEBPB | THP-1 | eGFP-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (37,938) | |
| CEBPB | THP-1 | NS1-Pam3csk-0h | GEO | Homo sapiens | GSE103477 | Download (15,146) | |
| CEBPB | THP-1 | NS1-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (32,456) | |
| CEBPB | WA01 | ENCODE | Homo sapiens | ENCSR000EBV | Download (30,293) | ||
| CEBPB | HCT-116 | ENCODE | Homo sapiens | ENCSR000BSD | Download (17,586) | ||
| CEBPB | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EDA | Download (136,526) | ||
| CEBPB | OCI-AML3 | GEO | Homo sapiens | GSE104745 | Download (4,840) | ||
| CEBPB | OCI-AML3 | JQ1_500nM_24h | GEO | Homo sapiens | GSE104745 | Download (1,815) | |
| CEBPB | epididymis | HEE | GEO | Homo sapiens | GSE109061 | Download (80,900) | |
| CEBPB | epididymis | HEE_R1881 | GEO | Homo sapiens | GSE109061 | Download (90,283) | |
| CEBPD | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BQJ | Download (35,741) | ||
| CEBPD | K-562 | ENCODE | Homo sapiens | ENCSR000BVY | Download (33,480) | ||
| CEBPD | HAEC | IL1b_4h | GEO | Homo sapiens | GSE89970 | Download (28,410) | |
| CEBPG | K-562 | ENCODE | Homo sapiens | ENCSR620VIC | Download (29,648) | ||
| CEBPG | MCF-7 | ENCODE | Homo sapiens | ENCSR094ZCF | Download (4,807) | ||
| CEBPG | Hep-G2 | ENCODE | Homo sapiens | ENCSR639IIZ | Download (111,819) | ||
| CEBPG | K-562 | ENCODE | Homo sapiens | ENCSR490LWA | Download (47,867) | ||
| CEBPZ | GM12878 | ENCODE | Homo sapiens | ENCSR347NOB | Download (2,406) | ||
| CEBPZ | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EDO | Download (288) | ||
| CEBPZ | K-562 | ENCODE | Homo sapiens | ENCSR618GDK | Download (1,443) | ||
| CHAMP1 | K-562 | ENCODE | Homo sapiens | ENCSR065XVO | Download (11,092) | ||
| CHAMP1 | K-562 | ENCODE | Homo sapiens | ENCSR315NNL | Download (2,684) | ||
| CHD1 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZE | Download (618) | ||
| CHD1 | A-549 | ENCODE | Homo sapiens | ENCSR398YBM | Download (20,480) | ||
| CHD1 | IMR-90 | ENCODE | Homo sapiens | ENCSR000EFC | Download (26,051) | ||
| CHD1 | K-562 | ENCODE | Homo sapiens | ENCSR000AQD | Download (15,216) | ||
| CHD1 | LNCaP | GEO | Homo sapiens | GSE64528 | Download (5,891) | ||
| CHD1 | LNCaP | DHT | GEO | Homo sapiens | GSE64528 | Download (5,825) | |
| CHD1 | LNCaP | DHT_KDCHD1 | GEO | Homo sapiens | GSE64528 | Download (8) | |
| CHD1 | hMSC-TERT | GEO | Homo sapiens | GSE89179 | Download (39,469) | ||
| CHD1 | hMSC-TERT | adipocyte | GEO | Homo sapiens | GSE89179 | Download (3,250) | |
| CHD1 | hMSC-TERT | osteoblast | GEO | Homo sapiens | GSE89179 | Download (29,071) | |
| CHD1 | MCF-7 | ENCODE | Homo sapiens | ENCSR360JOC | Download (14,668) | ||
| CHD1 | WA01 | ENCODE | Homo sapiens | ENCSR000AQK | Download (36,704) | ||
| CHD1 | WA01 | ENCODE | Homo sapiens | ENCSR000EBU | Download (5,409) | ||
| CHD1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR392EAA | Download (40,141) | ||
| CHD2 | SK-N-SH | ENCODE | Homo sapiens | ENCSR274SLQ | Download (57,236) | ||
| CHD2 | WA01 | ENCODE | Homo sapiens | ENCSR000EBT | Download (18,440) | ||
| CHD2 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECP | Download (45,818) | ||
| CHD2 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZR | Download (8,645) | ||
| CHD2 | A-549 | ENCODE | Homo sapiens | ENCSR067HGI | Download (6,735) | ||
| CHD2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EED | Download (17,681) | ||
| CHD2 | K-562 | ENCODE | Homo sapiens | ENCSR000EHD | Download (21,374) | ||
| CHD4 | GM12878 | ENCODE | Homo sapiens | ENCSR751CJG | Download (5,188) | ||
| CHD4 | A-549 | ENCODE | Homo sapiens | ENCSR550SCU | Download (14,707) | ||
| CHD4 | K-562 | ENCODE | Homo sapiens | ENCSR000ATL | Download (887) | ||
| CHD4 | SCC-9 | GEO | Homo sapiens | GSE97839 | Download (3,631) | ||
| CHD4 | SCC-9 | DOC1 | GEO | Homo sapiens | GSE97839 | Download (5,808) | |
| CHD7 | K-562 | ENCODE | Homo sapiens | ENCSR000AVD | Download (2,492) | ||
| CHD7 | WA01 | ENCODE | Homo sapiens | ENCSR000AVA | Download (16,734) | ||
| CHD7 | hiPSC | GEO | Homo sapiens | GSE108506 | Download (100,805) | ||
| CHD7 | hiPSC | AF22_abD3F5 | GEO | Homo sapiens | GSE108506 | Download (7,307) | |
| CHD7 | hiPSC | derived_lt-NES | GEO | Homo sapiens | GSE108506 | Download (62,308) | |
| CHD7 | hiPSC | derived_lt-NES_AF22 | GEO | Homo sapiens | GSE108506 | Download (259) | |
| CHD7 | hiPSC | derived_neural-crest-cell | GEO | Homo sapiens | GSE108506 | Download (28,628) | |
| CHD8 | HNSC | GEO | Homo sapiens | GSE57369 | Download (270,363) | ||
| CHD8 | T-47D | GEO | Homo sapiens | GSE62428 | Download (22,464) | ||
| CHD8 | T-47D | ETOH_45 | GEO | Homo sapiens | GSE62428 | Download (22,957) | |
| CHD8 | T-47D | ETOH_5 | GEO | Homo sapiens | GSE62428 | Download (11,881) | |
| CHD8 | T-47D | R5020_45 | GEO | Homo sapiens | GSE62428 | Download (8,271) | |
| CHD8 | T-47D | R5020_5 | GEO | Homo sapiens | GSE62428 | Download (2,809) | |
| CHR27 | Col-0 | Col-0_seedling | 6d-ACC | GEO | Arabidopsis thaliana | GSE79453 | Download (728) |
| CHR27 | Col-0 | Col-0_seedling | 6d-NHS | GEO | Arabidopsis thaliana | GSE79453 | Download (722) |
| CIITA | Raji | AB1 | GEO | Homo sapiens | GSE52941 | Download (1,552) | |
| CLF | Col-0 | Col-0_seedling | 10d | GEO | Arabidopsis thaliana | GSE108960 | Download (1,913) |
| CLOCK | neural | progenitor-O62409 | GEO | Homo sapiens | GSE96659 | Download (408) | |
| CLOCK | BA10-neocortex | 0 | GEO | Homo sapiens | GSE96659 | Download (667) | |
| CLOCK | BA10-neocortex | 1 | GEO | Homo sapiens | GSE96659 | Download (1,837) | |
| CLOCK | BA10-neocortex | 2 | GEO | Homo sapiens | GSE96659 | Download (11,458) | |
| CLOCK | BA10-neocortex | 3 | GEO | Homo sapiens | GSE96659 | Download (1,107) | |
| CLOCK | BA10-neocortex | 4 | GEO | Homo sapiens | GSE96659 | Download (2,027) | |
| CLOCK | BA40-neocortex | 0 | GEO | Homo sapiens | GSE96659 | Download (11,399) | |
| CLOCK | BA40-neocortex | 1 | GEO | Homo sapiens | GSE96659 | Download (981) | |
| CLOCK | BA40-neocortex | 2 | GEO | Homo sapiens | GSE96659 | Download (1,076) | |
| CLOCK | BA40-neocortex | 3 | GEO | Homo sapiens | GSE96659 | Download (14,295) | |
| CLOCK | BA40-neocortex | 4 | GEO | Homo sapiens | GSE96659 | Download (1,763) | |
| CLOCK | U2OS | GEO | Homo sapiens | GSE44236 | Download (24,360) | ||
| CLOCK | MCF-7 | ENCODE | Homo sapiens | ENCSR699YFX | Download (10,884) | ||
| CLOCK | MCF-7 | ENCODE | Homo sapiens | ENCSR934JDG | Download (10,151) | ||
| COBLL1 | LTAD | EtOH | GEO | Homo sapiens | GSE94577 | Download (1,431) | |
| CREB1 | MCF-7 | ENCODE | Homo sapiens | ENCSR620DUQ | Download (76,716) | ||
| CREB1 | MCF-7 | ENCODE | Homo sapiens | ENCSR897JAS | Download (78,357) | ||
| CREB1 | WA01 | ENCODE | Homo sapiens | ENCSR000BSN | Download (50,535) | ||
| CREB1 | MDA-MB-134 | GEO | Homo sapiens | GSE109103 | Download (15,061) | ||
| CREB1 | MDA-MB-134 | E2 | GEO | Homo sapiens | GSE109103 | Download (2,215) | |
| CREB1 | MDA-MB-134 | FI | GEO | Homo sapiens | GSE109103 | Download (13,668) | |
| CREB1 | GM12878 | ENCODE | Homo sapiens | ENCSR000BUF | Download (41,629) | ||
| CREB1 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BUR | Download (6,870) | ||
| CREB1 | A-549 | ENCODE | Homo sapiens | ENCSR000BRA | Download (7,553) | ||
| CREB1 | A-549 | ENCODE | Homo sapiens | ENCSR000BRB | Download (52,350) | ||
| CREB1 | A-549 | ENCODE | Homo sapiens | ENCSR000BRC | Download (21,795) | ||
| CREB1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BVL | Download (31,800) | ||
| CREB1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR112ALD | Download (60,266) | ||
| CREB1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR331ORD | Download (5,810) | ||
| CREB1 | K-562 | ENCODE | Homo sapiens | ENCSR000BSO | Download (24,568) | ||
| CREB1 | LNCaP | GEO | Homo sapiens | GSE63034 | Download (25,293) | ||
| CREB1 | Hep-G2 | GEO | Homo sapiens | GSE72082 | Download (81,929) | ||
| CREB1 | KG-1 | DMSO | GEO | Homo sapiens | GSE74928 | Download (481) | |
| CREB1 | KG-1 | XX65023 | GEO | Homo sapiens | GSE74928 | Download (5,378) | |
| CREB1 | LNCaP-abl | GEO | Homo sapiens | GSE63034 | Download (27,249) | ||
| CREB1 | LNCaP-abl | SHCTR | GEO | Homo sapiens | GSE63034 | Download (52,281) | |
| CREB1 | LNCaP-abl | SHFOXA1 | GEO | Homo sapiens | GSE63034 | Download (59,176) | |
| CREB3 | K-562 | ENCODE | Homo sapiens | ENCSR093FKD | Download (12,751) | ||
| CREB3L1 | K-562 | ENCODE | Homo sapiens | ENCSR109YGM | Download (14,682) | ||
| CREBBP | K-562 | ENCODE | Homo sapiens | ENCSR000ATT | Download (2,417) | ||
| CREBBP | MCF-7 | ENA | Homo sapiens | ERP000901 | Download (68,346) | ||
| CREBBP | MCF-7 | E2 | ENA | Homo sapiens | ERP000901 | Download (99,235) | |
| CREBBP | H3396 | E2 | GEO | Homo sapiens | GSE32349 | Download (2,833) | |
| CREBBP | H3396 | E2 | GEO | Homo sapiens | GSE32349 | Download (19,221) | |
| CREBBP | H3396 | ETOH | GEO | Homo sapiens | GSE32349 | Download (8,307) | |
| CREBBP | H3396 | GEO | Homo sapiens | GSE32349 | Download (2,860) | ||
| CREBBP | tonsil | GCBc_p4 | GEO | Homo sapiens | GSE89688 | Download (31,915) | |
| CREBBP | tonsil | GCBC_p5 | GEO | Homo sapiens | GSE89688 | Download (25,386) | |
| CREBBP | LS180 | GEO | Homo sapiens | GSE39277 | Download (8,149) | ||
| CREBBP | LS180 | 125 | GEO | Homo sapiens | GSE39277 | Download (33,063) | |
| CREM | GM12878 | ENCODE | Homo sapiens | ENCSR839XZU | Download (46,157) | ||
| CREM | Hep-G2 | ENCODE | Homo sapiens | ENCSR903ELW | Download (27,359) | ||
| CREM | K-562 | ENCODE | Homo sapiens | ENCSR077DKV | Download (65,678) | ||
| CRF4 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (1,502) |
| CRY1 | U2OS | GEO | Homo sapiens | GSE44236 | Download (23,589) | ||
| CRY2 | Col-0 | Col-0_seedling | 5d | GEO | Arabidopsis thaliana | GSE68193 | Download (2,572) |
| CSNK2A1 | LNCaP | DHT | GEO | Homo sapiens | GSE58607 | Download (15,848) | |
| CTBP1 | LNCaP | DHT1H | GEO | Homo sapiens | GSE58428 | Download (114) | |
| CTBP1 | MCF-7 | ENCODE | Homo sapiens | ENCSR636EYA | Download (46,407) | ||
| CTBP1 | HEK293T | ENCODE | Homo sapiens | ENCSR237TFX | Download (4,284) | ||
| CTBP1 | K-562 | ENCODE | Homo sapiens | ENCSR201NQZ | Download (72,620) | ||
| CTBP2 | LNCaP | DHT24H | GEO | Homo sapiens | GSE58428 | Download (4,203) | |
| CTBP2 | MCF-7 | GEO | Homo sapiens | GSE107013 | Download (21,845) | ||
| CTBP2 | MCF-7 | trps1_ko | GEO | Homo sapiens | GSE107013 | Download (4,726) | |
| CTBP2 | WA01 | ENCODE | Homo sapiens | ENCSR000EUO | Download (28,227) | ||
| CTCF | MCF-10A | ENA | Homo sapiens | ERP000783 | Download (34,789) | ||
| CTCF | MCF-7 | ENCODE | Homo sapiens | ENCSR000AHD | Download (59,708) | ||
| CTCF | MCF-7 | ENCODE | Homo sapiens | ENCSR000DML | Download (86,705) | ||
| CTCF | MCF-7 | ENCODE | Homo sapiens | ENCSR000DMO | Download (118,477) | ||
| CTCF | MCF-7 | ENCODE | Homo sapiens | ENCSR000DMR | Download (75,222) | ||
| CTCF | MCF-7 | ENCODE | Homo sapiens | ENCSR000DMS | Download (54,555) | ||
| CTCF | MCF-7 | ENCODE | Homo sapiens | ENCSR000DMV | Download (101,702) | ||
| CTCF | MCF-7 | ENCODE | Homo sapiens | ENCSR000DWH | Download (65,362) | ||
| CTCF | MCF-7 | ENCODE | Homo sapiens | ENCSR560BUE | Download (79,368) | ||
| CTCF | MCF-7 | E2_SHCTCF | ENA | Homo sapiens | ERP000209 | Download (37,736) | |
| CTCF | MCF-7 | E2_SHCTR | ENA | Homo sapiens | ERP000209 | Download (89,280) | |
| CTCF | MCF-7 | ENA | Homo sapiens | ERP000380 | Download (49,533) | ||
| CTCF | MCF-7 | E2 | ENA | Homo sapiens | ERP000380 | Download (55,070) | |
| CTCF | MCF-7 | TAM | ENA | Homo sapiens | ERP000380 | Download (52,770) | |
| CTCF | MCF-7 | TAM | ENA | Homo sapiens | ERP000783 | Download (46,913) | |
| CTCF | MCF-7 | GEO | Homo sapiens | GSE107749 | Download (86,154) | ||
| CTCF | MCF-7 | estrogen_45min | GEO | Homo sapiens | GSE107749 | Download (80,530) | |
| CTCF | NB69 | GEO | Homo sapiens | GSE101295 | Download (72,369) | ||
| CTCF | NCI-H929 | ENCODE | Homo sapiens | ENCSR634OAQ | Download (52,064) | ||
| CTCF | OCI-Ly1 | ENCODE | Homo sapiens | ENCSR072EUE | Download (67,103) | ||
| CTCF | OCI-Ly3 | ENCODE | Homo sapiens | ENCSR756ZKG | Download (77,452) | ||
| CTCF | OCI-Ly7 | ENCODE | Homo sapiens | ENCSR027HML | Download (58,825) | ||
| CTCF | PANC-1 | ENCODE | Homo sapiens | ENCSR203QEB | Download (38,645) | ||
| CTCF | PC-3 | ENCODE | Homo sapiens | ENCSR359LOD | Download (76,996) | ||
| CTCF | RWPE-1 | ENCODE | Homo sapiens | ENCSR303GFI | Download (32,768) | ||
| CTCF | RWPE-2 | ENCODE | Homo sapiens | ENCSR856JJB | Download (63,076) | ||
| CTCF | SH-SY5Y | GEO | Homo sapiens | GSE101295 | Download (57,523) | ||
| CTCF | SK-N-AS | GEO | Homo sapiens | GSE101295 | Download (63,238) | ||
| CTCF | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BLX | Download (54,518) | ||
| CTCF | SK-N-SH | ENCODE | Homo sapiens | ENCSR000DXQ | Download (67,983) | ||
| CTCF | SK-N-SH | ENCODE | Homo sapiens | ENCSR000EIC | Download (52,670) | ||
| CTCF | SK-N-SH | ENCODE | Homo sapiens | ENCSR541AMF | Download (66,569) | ||
| CTCF | SU-DHL-6 | ENCODE | Homo sapiens | ENCSR125DKL | Download (44,527) | ||
| CTCF | T-47D | ENCODE | Homo sapiens | ENCSR000BNO | Download (29,379) | ||
| CTCF | THP-1 | eGFP-IFNb-r1 | GEO | Homo sapiens | GSE103477 | Download (26,579) | |
| CTCF | THP-1 | Pam3csk-000m-Flavo-000m | GEO | Homo sapiens | GSE103477 | Download (64,229) | |
| CTCF | THP-1 | Pam3csk-000m-Flavo-240m | GEO | Homo sapiens | GSE103477 | Download (62,579) | |
| CTCF | THP-1 | Pam3csk-020m-Flavo-000m | GEO | Homo sapiens | GSE103477 | Download (60,846) | |
| CTCF | THP-1 | Pam3csk-025m-Flavo-000m | GEO | Homo sapiens | GSE103477 | Download (64,779) | |
| CTCF | THP-1 | Pam3csk-030m-Flavo-000m | GEO | Homo sapiens | GSE103477 | Download (60,239) | |
| CTCF | THP-1 | Pam3csk-045m-Flavo-000m | GEO | Homo sapiens | GSE103477 | Download (62,938) | |
| CTCF | THP-1 | Pam3csk-060m-Flavo-000m | GEO | Homo sapiens | GSE103477 | Download (60,023) | |
| CTCF | THP-1 | Pam3csk-120m-Flavo-000m | GEO | Homo sapiens | GSE103477 | Download (53,278) | |
| CTCF | THP-1 | Pam3csk-150m-Flavo-030m | GEO | Homo sapiens | GSE103477 | Download (63,837) | |
| CTCF | THP-1 | Pam3csk-180m-Flavo-060m | GEO | Homo sapiens | GSE103477 | Download (63,405) | |
| CTCF | THP-1 | Pam3csk-360m-Flavo-240m | GEO | Homo sapiens | GSE103477 | Download (63,890) | |
| CTCF | THP-1 | PMA_Dex-0h | GEO | Homo sapiens | GSE103477 | Download (31,561) | |
| CTCF | THP-1 | PMA_Dex-6h | GEO | Homo sapiens | GSE103477 | Download (41,906) | |
| CTCF | THP-1 | siCtrl-eGFP-Pam3csk-0h | GEO | Homo sapiens | GSE103477 | Download (50,391) | |
| CTCF | THP-1 | siCtrl-eGFP-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (46,829) | |
| CTCF | THP-1 | siCtrl-NS1-Pam3csk-0h | GEO | Homo sapiens | GSE103477 | Download (47,895) | |
| CTCF | THP-1 | siCtrl-NS1-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (47,409) | |
| CTCF | THP-1 | siCtrl-NS1-Pam3csk-7h-Flavo-3h | GEO | Homo sapiens | GSE103477 | Download (40,821) | |
| CTCF | THP-1 | siNIPBL-eGFP-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (43,306) | |
| CTCF | THP-1 | siNIPBL-NS1-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (44,132) | |
| CTCF | THP-1 | siNIPBL-NS1-Pam3csk-7h-Flavo-3h | GEO | Homo sapiens | GSE103477 | Download (47,530) | |
| CTCF | THP-1 | siWAPL-eGFP-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (47,692) | |
| CTCF | THP-1 | siWAPL-NS1-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (49,965) | |
| CTCF | THP-1 | siWAPL-NS1-Pam3csk-7h-Flavo-3h | GEO | Homo sapiens | GSE103477 | Download (44,480) | |
| CTCF | U-937 | ENA | Homo sapiens | ERP008568 | Download (13,522) | ||
| CTCF | WA01 | ENCODE | Homo sapiens | ENCSR000AMF | Download (85,840) | ||
| CTCF | WA01 | ENCODE | Homo sapiens | ENCSR000BNH | Download (49,862) | ||
| CTCF | WA01 | ENCODE | Homo sapiens | ENCSR000DLK | Download (53,628) | ||
| CTCF | WERI-Rb-1 | ENCODE | Homo sapiens | ENCSR000DXW | Download (66,724) | ||
| CTCF | WI-38 | ENCODE | Homo sapiens | ENCSR000DYB | Download (30,691) | ||
| CTCF | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000AOA | Download (67,356) | ||
| CTCF | B-cell | ENCODE | Homo sapiens | ENCSR000AUV | Download (47,900) | ||
| CTCF | HCT-116 | ENCODE | Homo sapiens | ENCSR000BSE | Download (40,168) | ||
| CTCF | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000DLO | Download (58,464) | ||
| CTCF | cardiac | muscle | ENCODE | Homo sapiens | ENCSR000DTI | Download (53,937) | |
| CTCF | choroid-plexus | epithelial | ENCODE | Homo sapiens | ENCSR000DTL | Download (66,146) | |
| CTCF | HCT-116 | ENCODE | Homo sapiens | ENCSR000DTO | Download (52,432) | ||
| CTCF | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000DUB | Download (60,224) | ||
| CTCF | bonchial | epithelial | ENCODE | Homo sapiens | ENCSR000DXI | Download (69,009) | |
| CTCF | esophagus | squamous-epithelium | ENCODE | Homo sapiens | ENCSR003SZZ | Download (32,841) | |
| CTCF | adrenal-gland | ENCODE | Homo sapiens | ENCSR014GSQ | Download (28,662) | ||
| CTCF | skin | suprapubic | ENCODE | Homo sapiens | ENCSR038FOS | Download (6,592) | |
| CTCF | neuron | bipolar_doxy_4d | ENCODE | Homo sapiens | ENCSR066BZZ | Download (82,913) | |
| CTCF | cardiac | right-atrium-auricular-region | ENCODE | Homo sapiens | ENCSR066GBX | Download (22,245) | |
| CTCF | gastrocnemius-medialis | ENCODE | Homo sapiens | ENCSR071XWO | Download (25,888) | ||
| CTCF | esophagus | muscularis-mucosa | ENCODE | Homo sapiens | ENCSR074SFL | Download (29,899) | |
| CTCF | omental-fat-pad | ENCODE | Homo sapiens | ENCSR089DTY | Download (4,041) | ||
| CTCF | thyroid-gland | ENCODE | Homo sapiens | ENCSR145HBC | Download (44,659) | ||
| CTCF | gastroesophageal-sphincter | ENCODE | Homo sapiens | ENCSR146BGM | Download (47,007) | ||
| CTCF | stomach | ENCODE | Homo sapiens | ENCSR173AIR | Download (24,042) | ||
| CTCF | coronary-artery | ENCODE | Homo sapiens | ENCSR175FLL | Download (39,118) | ||
| CTCF | gastroesophageal-sphincter | ENCODE | Homo sapiens | ENCSR186NVR | Download (47,697) | ||
| CTCF | gastroesophageal-sphincter | ENCODE | Homo sapiens | ENCSR206ETG | Download (41,331) | ||
| CTCF | omental-fat-pad | ENCODE | Homo sapiens | ENCSR225OKX | Download (11,864) | ||
| CTCF | HCT-116 | ENCODE | Homo sapiens | ENCSR240PRQ | Download (70,426) | ||
| CTCF | skin | lower-leg | ENCODE | Homo sapiens | ENCSR252XWG | Download (18,894) | |
| CTCF | smooth-muscle-cell | ENCODE | Homo sapiens | ENCSR261VAS | Download (48,472) | ||
| CTCF | esophagus | muscularis-mucosa | ENCODE | Homo sapiens | ENCSR353DFU | Download (38,213) | |
| CTCF | stomach | ENCODE | Homo sapiens | ENCSR361KVZ | Download (30,464) | ||
| CTCF | Peyers-patch | ENCODE | Homo sapiens | ENCSR375VXU | Download (52,548) | ||
| CTCF | Peyers-patch | ENCODE | Homo sapiens | ENCSR391ZKN | Download (23,162) | ||
| CTCF | uterus | ENCODE | Homo sapiens | ENCSR392SFJ | Download (36,496) | ||
| CTCF | MM1-S | ENCODE | Homo sapiens | ENCSR402IDP | Download (53,463) | ||
| CTCF | adrenal-gland | ENCODE | Homo sapiens | ENCSR408ZEE | Download (30,575) | ||
| CTCF | Peyers-patch | ENCODE | Homo sapiens | ENCSR419ANE | Download (44,207) | ||
| CTCF | gastrocnemius-medialis | ENCODE | Homo sapiens | ENCSR428BKN | Download (31,298) | ||
| CTCF | skin | lower-leg | ENCODE | Homo sapiens | ENCSR430TEE | Download (488) | |
| CTCF | nerve | tibial | ENCODE | Homo sapiens | ENCSR434XLP | Download (44,515) | |
| CTCF | adrenal-gland | ENCODE | Homo sapiens | ENCSR450BLH | Download (46,188) | ||
| CTCF | esophagus | squamous-epithelium | ENCODE | Homo sapiens | ENCSR450FRI | Download (58,398) | |
| CTCF | LNCaP-C4-2B | ENCODE | Homo sapiens | ENCSR460LGH | Download (51,115) | ||
| CTCF | nerve | tibial | ENCODE | Homo sapiens | ENCSR469POZ | Download (40,885) | |
| CTCF | thyroid-gland | ENCODE | Homo sapiens | ENCSR492ZIW | Download (39,716) | ||
| CTCF | ovary | ENCODE | Homo sapiens | ENCSR493APD | Download (20,482) | ||
| CTCF | testis | ENCODE | Homo sapiens | ENCSR494TNM | Download (22,399) | ||
| CTCF | skin | suprapubic | ENCODE | Homo sapiens | ENCSR515LRI | Download (4,984) | |
| CTCF | uterus | ENCODE | Homo sapiens | ENCSR527TPP | Download (36,272) | ||
| CTCF | heart | left-ventricle | ENCODE | Homo sapiens | ENCSR544APK | Download (58,701) | |
| CTCF | ovary | ENCODE | Homo sapiens | ENCSR548DDS | Download (21,646) | ||
| CTCF | stomach | ENCODE | Homo sapiens | ENCSR549WAU | Download (16,913) | ||
| CTCF | esophagus | muscularis-mucosa | ENCODE | Homo sapiens | ENCSR559KAB | Download (43,191) | |
| CTCF | skin | lower-leg | ENCODE | Homo sapiens | ENCSR582MTM | Download (4,341) | |
| CTCF | vagina | ENCODE | Homo sapiens | ENCSR606TNN | Download (26,234) | ||
| CTCF | stomach | ENCODE | Homo sapiens | ENCSR618QYE | Download (894) | ||
| CTCF | neuron | bipolar_doxy_4d | ENCODE | Homo sapiens | ENCSR619IUE | Download (74,941) | |
| CTCF | vagina | ENCODE | Homo sapiens | ENCSR655ECZ | Download (16,301) | ||
| CTCF | gastroesophageal-sphincter | ENCODE | Homo sapiens | ENCSR661XNQ | Download (37,556) | ||
| CTCF | aorta | thoracic | ENCODE | Homo sapiens | ENCSR668BTN | Download (44,390) | |
| CTCF | cardiac-muscle | ENCODE | Homo sapiens | ENCSR713SXF | Download (49,953) | ||
| CTCF | heart | left-ventricle | ENCODE | Homo sapiens | ENCSR718SDR | Download (49,282) | |
| CTCF | thyroid-gland | ENCODE | Homo sapiens | ENCSR744YJR | Download (27,040) | ||
| CTCF | skin | suprapubic | ENCODE | Homo sapiens | ENCSR756KRS | Download (9,676) | |
| CTCF | esophagus | squamous-epithelium | ENCODE | Homo sapiens | ENCSR756URL | Download (32,950) | |
| CTCF | subcutaneous-adipose-tissue | ENCODE | Homo sapiens | ENCSR762PCG | Download (485) | ||
| CTCF | adrenal-gland | ENCODE | Homo sapiens | ENCSR770IWO | Download (49,367) | ||
| CTCF | esophagus | squamous-epithelium | ENCODE | Homo sapiens | ENCSR773JBP | Download (39,886) | |
| CTCF | neutrophil | ENCODE | Homo sapiens | ENCSR785YRL | Download (33,303) | ||
| CTCF | heart | left-ventricle | ENCODE | Homo sapiens | ENCSR791AYW | Download (53,449) | |
| CTCF | uterus | ENCODE | Homo sapiens | ENCSR798NVH | Download (31,796) | ||
| CTCF | Peyers-patch | ENCODE | Homo sapiens | ENCSR799WDT | Download (42,296) | ||
| CTCF | nerve | tibial | ENCODE | Homo sapiens | ENCSR822PJT | Download (49,546) | |
| CTCF | esophagus | squamous-epithelium | ENCODE | Homo sapiens | ENCSR838RUX | Download (46,380) | |
| CTCF | adrenal-gland | ENCODE | Homo sapiens | ENCSR899JSO | Download (39,510) | ||
| CTCF | thyroid-gland | ENCODE | Homo sapiens | ENCSR955BIB | Download (38,433) | ||
| CTCF | aorta | ascending | ENCODE | Homo sapiens | ENCSR960MDF | Download (20,173) | |
| CTCF | testis | ENCODE | Homo sapiens | ENCSR981CID | Download (35,198) | ||
| CTCF | gastrocnemius-medialis | ENCODE | Homo sapiens | ENCSR998NQG | Download (20,074) | ||
| CTCF | 81-3 | ENA | Homo sapiens | ERP002246 | Download (33,071) | ||
| CTCF | 226LDM | GEO | Homo sapiens | GSE102237 | Download (3,849) | ||
| CTCF | HeLa-Kyoto | GEO | Homo sapiens | GSE102884 | Download (47,657) | ||
| CTCF | HeLa-Kyoto | PDS5-depleted | GEO | Homo sapiens | GSE102884 | Download (59,012) | |
| CTCF | HeLa-Kyoto | WAPL-depleted | GEO | Homo sapiens | GSE102884 | Download (61,010) | |
| CTCF | HeLa-Kyoto | WAPL_PDS-depleted | GEO | Homo sapiens | GSE102884 | Download (54,435) | |
| CTCF | MDM | GEO | Homo sapiens | GSE103477 | Download (30,179) | ||
| CTCF | MDM | dNS1 | GEO | Homo sapiens | GSE103477 | Download (30,413) | |
| CTCF | MDM | H5N1 | GEO | Homo sapiens | GSE103477 | Download (31,546) | |
| CTCF | MDM | IFNb | GEO | Homo sapiens | GSE103477 | Download (28,868) | |
| CTCF | HCT-116 | RAD21-mAC | GEO | Homo sapiens | GSE104888 | Download (54,358) | |
| CTCF | HCT-116 | RAD21-mAC_500uM_auxin | GEO | Homo sapiens | GSE104888 | Download (41,459) | |
| CTCF | HSPC-CD34 | GEO | Homo sapiens | GSE107147 | Download (42,229) | ||
| CTCF | HeLa-S3 | synchro | GEO | Homo sapiens | GSE108173 | Download (28,205) | |
| CTCF | HeLa-S3 | unsynchro | GEO | Homo sapiens | GSE108173 | Download (54,630) | |
| CTCF | ASC | GEO | Homo sapiens | GSE21366 | Download (38,995) | ||
| CTCF | BJAB | GEO | Homo sapiens | GSE31485 | Download (25,403) | ||
| CTCF | BL41 | GEO | Homo sapiens | GSE31485 | Download (30,031) | ||
| CTCF | erythroid | GEO | Homo sapiens | GSE67783 | Download (59,907) | ||
| CTCF | GM12878 | GEO | Homo sapiens | GSE76922 | Download (43,742) | ||
| CTCF | GP5D | GEO | Homo sapiens | GSE51234 | Download (52,813) | ||
| CTCF | GP5D | SIRAD21 | GEO | Homo sapiens | GSE51234 | Download (29,624) | |
| CTCF | HEK293 | GEO | Homo sapiens | GSE68976 | Download (14,524) | ||
| CTCF | HEK293 | GEO | Homo sapiens | GSE76494 | Download (167,785) | ||
| CTCF | 22Rv1 | hydroxy_10nM_4h | ENCODE | Homo sapiens | ENCSR847XGE | Download (49,183) | |
| CTCF | 22Rv1 | ENCODE | Homo sapiens | ENCSR857PBV | Download (49,761) | ||
| CTCF | artery | tibial | ENCODE | Homo sapiens | ENCSR699BEK | Download (41,379) | |
| CTCF | astrocyte | ENCODE | Homo sapiens | ENCSR000AOO | Download (42,488) | ||
| CTCF | astrocyte | spinal_cord | ENCODE | Homo sapiens | ENCSR000DSU | Download (58,894) | |
| CTCF | astrocyte | cerebellum | ENCODE | Homo sapiens | ENCSR000DSZ | Download (53,405) | |
| CTCF | BE2C | ENCODE | Homo sapiens | ENCSR000DQD | Download (56,437) | ||
| CTCF | BJ | ENCODE | Homo sapiens | ENCSR000DQI | Download (53,073) | ||
| CTCF | breast | epithelium | ENCODE | Homo sapiens | ENCSR304XUZ | Download (20,719) | |
| CTCF | breast | epithelium | ENCODE | Homo sapiens | ENCSR661NXJ | Download (38,537) | |
| CTCF | breast | epithelium | ENCODE | Homo sapiens | ENCSR697YIN | Download (37,848) | |
| CTCF | CACO2 | ENCODE | Homo sapiens | ENCSR000DQN | Download (46,068) | ||
| CTCF | CD14 | ENCODE | Homo sapiens | ENCSR000ATN | Download (43,336) | ||
| CTCF | CHRF28811 | ENA | Homo sapiens | ERP008568 | Download (24,855) | ||
| CTCF | colon | transverse | ENCODE | Homo sapiens | ENCSR102CSD | Download (47,255) | |
| CTCF | colon | sigmoid | ENCODE | Homo sapiens | ENCSR222SQE | Download (19,019) | |
| CTCF | colon | transverse | ENCODE | Homo sapiens | ENCSR236YGF | Download (38,167) | |
| CTCF | colon | transverse | ENCODE | Homo sapiens | ENCSR449SEF | Download (18,882) | |
| CTCF | colon | transverse | ENCODE | Homo sapiens | ENCSR558HTE | Download (4,212) | |
| CTCF | colon | transverse | ENCODE | Homo sapiens | ENCSR608WPS | Download (31,805) | |
| CTCF | colon | sigmoid | ENCODE | Homo sapiens | ENCSR721AHD | Download (28,698) | |
| CTCF | colon | transverse | ENCODE | Homo sapiens | ENCSR769WKR | Download (33,762) | |
| CTCF | colon | transverse | ENCODE | Homo sapiens | ENCSR833FWC | Download (45,661) | |
| CTCF | colon | transverse | ENCODE | Homo sapiens | ENCSR907BES | Download (25,354) | |
| CTCF | colon | sigmoid | ENCODE | Homo sapiens | ENCSR925GDS | Download (12,220) | |
| CTCF | DND41 | ENCODE | Homo sapiens | ENCSR000AQU | Download (73,489) | ||
| CTCF | DOHH2 | ENCODE | Homo sapiens | ENCSR084RDK | Download (69,677) | ||
| CTCF | endothelial | umbilical-vein | ENCODE | Homo sapiens | ENCSR000ALA | Download (43,329) | |
| CTCF | endothelial | umbilical-vein | ENCODE | Homo sapiens | ENCSR000DLW | Download (44,823) | |
| CTCF | endothelial | brain-microvascular | ENCODE | Homo sapiens | ENCSR000DTA | Download (72,694) | |
| CTCF | endothelial | umbilical-vein | ENCODE | Homo sapiens | ENCSR000DVP | Download (57,113) | |
| CTCF | epithelial | mammary | ENCODE | Homo sapiens | ENCSR000ALV | Download (39,920) | |
| CTCF | epithelial | esophagus | ENCODE | Homo sapiens | ENCSR000DTR | Download (43,756) | |
| CTCF | epithelial | mammary | ENCODE | Homo sapiens | ENCSR000DUS | Download (54,918) | |
| CTCF | epithelial | kidney | ENCODE | Homo sapiens | ENCSR000DVH | Download (65,522) | |
| CTCF | epithelial | retinal_pigment | ENCODE | Homo sapiens | ENCSR000DVI | Download (53,606) | |
| CTCF | epithelial | proximal_tubule | ENCODE | Homo sapiens | ENCSR000DXD | Download (64,708) | |
| CTCF | erythroblast | adult_erythroblasts | GEO | Homo sapiens | GSE102184 | Download (63,294) | |
| CTCF | fetal | erythroblasts | GEO | Homo sapiens | GSE102184 | Download (49,969) | |
| CTCF | fibroblast | LUNG | ENCODE | Homo sapiens | ENCSR000ANO | Download (41,948) | |
| CTCF | fibroblast | DERMAL | ENCODE | Homo sapiens | ENCSR000APM | Download (43,589) | |
| CTCF | fibroblast | SKIN_LEG | ENCODE | Homo sapiens | ENCSR000DPG | Download (59,761) | |
| CTCF | fibroblast | LUNG | ENCODE | Homo sapiens | ENCSR000DPM | Download (47,118) | |
| CTCF | fibroblast | PEDAL_DIGIT_SKIN | ENCODE | Homo sapiens | ENCSR000DPP | Download (34,815) | |
| CTCF | fibroblast | GINGIVA | ENCODE | Homo sapiens | ENCSR000DPS | Download (55,544) | |
| CTCF | fibroblast | SKIN_ABDOMEN | ENCODE | Homo sapiens | ENCSR000DPV | Download (46,107) | |
| CTCF | fibroblast | AORTIC_ADVENTITIA | ENCODE | Homo sapiens | ENCSR000DPY | Download (51,812) | |
| CTCF | fibroblast | CARDIAC | ENCODE | Homo sapiens | ENCSR000DTF | Download (59,812) | |
| CTCF | fibroblast | FORESKIN | ENCODE | Homo sapiens | ENCSR000DUH | Download (48,869) | |
| CTCF | fibroblast | MAMMARY | ENCODE | Homo sapiens | ENCSR000DUU | Download (43,816) | |
| CTCF | fibroblast | PULMONARY_ARTERY | ENCODE | Homo sapiens | ENCSR000DUX | Download (56,199) | |
| CTCF | fibroblast | LUNG | ENCODE | Homo sapiens | ENCSR000DVA | Download (42,823) | |
| CTCF | fibroblast | VILLOUS_MESENCHYME | ENCODE | Homo sapiens | ENCSR000DVQ | Download (55,893) | |
| CTCF | fibroblast | FORESKIN | ENCODE | Homo sapiens | ENCSR000DWQ | Download (49,877) | |
| CTCF | fibroblast | LUNG | ENCODE | Homo sapiens | ENCSR000DWY | Download (44,314) | |
| CTCF | GM06990 | ENCODE | Homo sapiens | ENCSR000DQW | Download (50,480) | ||
| CTCF | GM10248 | ENCODE | Homo sapiens | ENCSR000DKP | Download (19,104) | ||
| CTCF | GM10266 | ENCODE | Homo sapiens | ENCSR000DKR | Download (28,081) | ||
| CTCF | GM12801 | ENCODE | Homo sapiens | ENCSR000DQY | Download (22,885) | ||
| CTCF | GM12864 | ENCODE | Homo sapiens | ENCSR000DRB | Download (43,554) | ||
| CTCF | GM12865 | ENCODE | Homo sapiens | ENCSR000DRE | Download (40,328) | ||
| CTCF | GM12866 | ENCODE | Homo sapiens | ENCSR000DRF | Download (56,077) | ||
| CTCF | GM12867 | ENCODE | Homo sapiens | ENCSR000DRH | Download (50,097) | ||
| CTCF | GM12868 | ENCODE | Homo sapiens | ENCSR000DRI | Download (52,699) | ||
| CTCF | GM12869 | ENCODE | Homo sapiens | ENCSR000DRJ | Download (59,921) | ||
| CTCF | GM12870 | ENCODE | Homo sapiens | ENCSR000DRK | Download (49,157) | ||
| CTCF | GM12872 | ENCODE | Homo sapiens | ENCSR000DRN | Download (41,811) | ||
| CTCF | GM12873 | ENCODE | Homo sapiens | ENCSR000DRP | Download (49,968) | ||
| CTCF | GM12874 | ENCODE | Homo sapiens | ENCSR000DRR | Download (49,404) | ||
| CTCF | GM12875 | ENCODE | Homo sapiens | ENCSR000DRU | Download (46,377) | ||
| CTCF | GM12878 | ENCODE | Homo sapiens | ENCSR000AKB | Download (34,372) | ||
| CTCF | GM12878 | ENCODE | Homo sapiens | ENCSR000DKV | Download (54,504) | ||
| CTCF | GM12878 | ENCODE | Homo sapiens | ENCSR000DRZ | Download (42,365) | ||
| CTCF | GM12878 | ENCODE | Homo sapiens | ENCSR000DZN | Download (46,767) | ||
| CTCF | GM12878 | ENA | Homo sapiens | ERP002246 | Download (25,766) | ||
| CTCF | GM12891 | ENCODE | Homo sapiens | ENCSR000DKX | Download (121,765) | ||
| CTCF | GM12892 | ENCODE | Homo sapiens | ENCSR000DKY | Download (152,664) | ||
| CTCF | GM13976 | ENCODE | Homo sapiens | ENCSR000DKZ | Download (13,682) | ||
| CTCF | GM13977 | ENCODE | Homo sapiens | ENCSR000DLB | Download (41,638) | ||
| CTCF | GM19238 | ENCODE | Homo sapiens | ENCSR000DLD | Download (95,395) | ||
| CTCF | GM19239 | ENCODE | Homo sapiens | ENCSR000DLE | Download (123,495) | ||
| CTCF | GM19240 | ENCODE | Homo sapiens | ENCSR000DLF | Download (103,705) | ||
| CTCF | GM20000 | ENCODE | Homo sapiens | ENCSR000DLG | Download (27,909) | ||
| CTCF | H54 | ENCODE | Homo sapiens | ENCSR000DKN | Download (67,906) | ||
| CTCF | HEK293 | ENCODE | Homo sapiens | ENCSR000DTW | Download (66,246) | ||
| CTCF | HEK293 | ENCODE | Homo sapiens | ENCSR617IFZ | Download (45,490) | ||
| CTCF | HEK293T | GEO | Homo sapiens | GSE103651 | Download (39,314) | ||
| CTCF | HeLa | dC9Sun-D3A_CDCC85C | GEO | Homo sapiens | GSE107607 | Download (59,053) | |
| CTCF | HeLa | dC9Sun-D3A_MIR152 | GEO | Homo sapiens | GSE107607 | Download (59,965) | |
| CTCF | HeLa | dC9Sun-D3AMut_MIR152 | GEO | Homo sapiens | GSE107607 | Download (52,525) | |
| CTCF | HeLa | dC9Sun-D3A_SHB | GEO | Homo sapiens | GSE107607 | Download (54,351) | |
| CTCF | hepatocyte | ENCODE | Homo sapiens | ENCSR252QYR | Download (56,272) | ||
| CTCF | hepatocyte | ENA | Homo sapiens | ERP000395 | Download (65,496) | ||
| CTCF | HFF | ENCODE | Homo sapiens | ENCSR000DUM | Download (42,242) | ||
| CTCF | Ishikawa | ENCODE | Homo sapiens | ENCSR000BQE | Download (20,561) | ||
| CTCF | islet | ENA | Homo sapiens | ERP004003 | Download (63,252) | ||
| CTCF | KARPAS422 | ENCODE | Homo sapiens | ENCSR113REG | Download (59,667) | ||
| CTCF | keratinocyte | ENCODE | Homo sapiens | ENCSR000ALJ | Download (63,434) | ||
| CTCF | keratinocyte | ENCODE | Homo sapiens | ENCSR000DNC | Download (70,261) | ||
| CTCF | keratinocyte | ENCODE | Homo sapiens | ENCSR000DWX | Download (68,211) | ||
| CTCF | kidney | ENCODE | Homo sapiens | ENCSR000DMC | Download (27,523) | ||
| CTCF | liver | ENCODE | Homo sapiens | ENCSR254YRM | Download (37,956) | ||
| CTCF | liver | right-lobe-of | ENCODE | Homo sapiens | ENCSR911GFJ | Download (36,918) | |
| CTCF | LNCaP | ENCODE | Homo sapiens | ENCSR000DME | Download (35,086) | ||
| CTCF | LNCaP | ENCODE | Homo sapiens | ENCSR000DMF | Download (41,298) | ||
| CTCF | LNCaP | ENCODE | Homo sapiens | ENCSR315NAC | Download (44,451) | ||
| CTCF | LNCaP | hydroxy_10nM_4h | ENCODE | Homo sapiens | ENCSR673WZL | Download (52,841) | |
| CTCF | Loucy | ENCODE | Homo sapiens | ENCSR464DKE | Download (73,361) | ||
| CTCF | lung | ENCODE | Homo sapiens | ENCSR000DMH | Download (28,507) | ||
| CTCF | lung | left_upper-lobe | ENCODE | Homo sapiens | ENCSR027FSZ | Download (44,284) | |
| CTCF | lung | left_upper-lobe | ENCODE | Homo sapiens | ENCSR799TJD | Download (34,827) | |
| CTCF | lung | left_upper-lobe | ENCODE | Homo sapiens | ENCSR964BKO | Download (31,705) | |
| CTCF | lung | left_upper-lobe | ENCODE | Homo sapiens | ENCSR970UZD | Download (24,750) | |
| CTCF | lung | left_upper-lobe | ENCODE | Homo sapiens | ENCSR972LYL | Download (40,030) | |
| CTCF | medulloblastoma | ENCODE | Homo sapiens | ENCSR000DMY | Download (42,028) | ||
| CTCF | myoblast | skeletal_muscle | ENCODE | Homo sapiens | ENCSR000ANE | Download (53,123) | |
| CTCF | myotube | ENCODE | Homo sapiens | ENCSR000ANS | Download (55,058) | ||
| CTCF | NB4 | ENCODE | Homo sapiens | ENCSR000DWN | Download (50,232) | ||
| CTCF | neural | progenitor | ENCODE | Homo sapiens | ENCSR125NBL | Download (63,014) | |
| CTCF | neural | ENCODE | Homo sapiens | ENCSR822CEA | Download (51,998) | ||
| CTCF | osteoblast | ENCODE | Homo sapiens | ENCSR000APF | Download (63,780) | ||
| CTCF | pancreas | ENCODE | Homo sapiens | ENCSR000DND | Download (48,146) | ||
| CTCF | pancreas | body | ENCODE | Homo sapiens | ENCSR265PFQ | Download (43,336) | |
| CTCF | pancreas | body | ENCODE | Homo sapiens | ENCSR307PFP | Download (39,420) | |
| CTCF | pancreas | body | ENCODE | Homo sapiens | ENCSR408XTO | Download (50,101) | |
| CTCF | pancreas | body | ENCODE | Homo sapiens | ENCSR484DDO | Download (41,792) | |
| CTCF | pancreas | body | ENCODE | Homo sapiens | ENCSR572DUJ | Download (32,572) | |
| CTCF | prostate | epithelial | ENCODE | Homo sapiens | ENCSR196HOM | Download (70,905) | |
| CTCF | prostate | gland | ENCODE | Homo sapiens | ENCSR720USO | Download (52,458) | |
| CTCF | prostate | gland | ENCODE | Homo sapiens | ENCSR829HTO | Download (37,094) | |
| CTCF | spleen | ENCODE | Homo sapiens | ENCSR000DNI | Download (39,305) | ||
| CTCF | spleen | ENCODE | Homo sapiens | ENCSR028YEV | Download (31,108) | ||
| CTCF | spleen | ENCODE | Homo sapiens | ENCSR234HEM | Download (16,695) | ||
| CTCF | spleen | ENCODE | Homo sapiens | ENCSR482PMN | Download (32,770) | ||
| CTCF | spleen | ENCODE | Homo sapiens | ENCSR692ILH | Download (18,201) | ||
| CTCF | VCaP | ENCODE | Homo sapiens | ENCSR265ARE | Download (56,280) | ||
| CTCF | WA09 | GEO | Homo sapiens | GSE105028 | Download (14,172) | ||
| CTCF | WA09 | heat-shock | GEO | Homo sapiens | GSE105028 | Download (7,616) | |
| CTCF | ZR751 | ENA | Homo sapiens | ERP000783 | Download (43,582) | ||
| CTCF | A-673 | ENCODE | Homo sapiens | ENCSR611JJS | Download (66,058) | ||
| CTCF | A-549 | ENCODE | Homo sapiens | ENCSR000AUE | Download (57,883) | ||
| CTCF | A-549 | ENCODE | Homo sapiens | ENCSR000AUF | Download (69,918) | ||
| CTCF | A-549 | ENCODE | Homo sapiens | ENCSR000BHV | Download (37,231) | ||
| CTCF | A-549 | ENCODE | Homo sapiens | ENCSR000BHW | Download (37,605) | ||
| CTCF | A-549 | ENCODE | Homo sapiens | ENCSR000DNA | Download (55,336) | ||
| CTCF | A-549 | ENCODE | Homo sapiens | ENCSR000DPF | Download (66,068) | ||
| CTCF | A-549 | ENCODE | Homo sapiens | ENCSR000DYD | Download (46,736) | ||
| CTCF | GM12871 | ENCODE | Homo sapiens | ENCSR000DRL | Download (61,473) | ||
| CTCF | GM23338 | ENCODE | Homo sapiens | ENCSR932ZMX | Download (112,600) | ||
| CTCF | GM23338 | ENCODE | Homo sapiens | ENCSR987GXT | Download (83,254) | ||
| CTCF | Hep-G2 | ENCODE | Homo sapiens | ENCSR000AMA | Download (54,128) | ||
| CTCF | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BIE | Download (63,410) | ||
| CTCF | Hep-G2 | ENCODE | Homo sapiens | ENCSR000DLS | Download (57,779) | ||
| CTCF | Hep-G2 | ENCODE | Homo sapiens | ENCSR000DUG | Download (62,433) | ||
| CTCF | Hep-G2 | ENA | Homo sapiens | ERP000209 | Download (51,676) | ||
| CTCF | HL-60 | ENCODE | Homo sapiens | ENCSR000DUP | Download (36,487) | ||
| CTCF | HL-60 | ENA | Homo sapiens | ERP008568 | Download (36,093) | ||
| CTCF | HUDEP-2 | 30min | GEO | Homo sapiens | GSE104676 | Download (58,456) | |
| CTCF | IMR-90 | ENCODE | Homo sapiens | ENCSR000EFI | Download (53,013) | ||
| CTCF | K-562 | ENCODE | Homo sapiens | ENCSR000AKO | Download (57,456) | ||
| CTCF | K-562 | ENCODE | Homo sapiens | ENCSR000BPJ | Download (53,476) | ||
| CTCF | K-562 | ENCODE | Homo sapiens | ENCSR000DMA | Download (67,159) | ||
| CTCF | K-562 | ENCODE | Homo sapiens | ENCSR000DWE | Download (48,316) | ||
| CTCF | K-562 | ENCODE | Homo sapiens | ENCSR000EGM | Download (59,212) | ||
| CTCF | WI-38VA13 | GEO | Homo sapiens | GSE41048 | Download (21,595) | ||
| CTCF | MM1-S | GEO | Homo sapiens | GSE43743 | Download (61,726) | ||
| CTCF | COLO-829 | GEO | Homo sapiens | GSE81945 | Download (38,327) | ||
| CTCF | primary-epidermal-keratinocyte | diff_d0 | GEO | Homo sapiens | GSE84657 | Download (70,634) | |
| CTCF | primary-epidermal-keratinocyte | diff_d3 | GEO | Homo sapiens | GSE84657 | Download (44,707) | |
| CTCF | primary-epidermal-keratinocyte | diff_d6 | GEO | Homo sapiens | GSE84657 | Download (51,847) | |
| CTCF | retina | AB1-FW14 | GEO | Homo sapiens | GSE86981 | Download (31,274) | |
| CTCF | retina | AB1-FW16 | GEO | Homo sapiens | GSE86981 | Download (36,131) | |
| CTCF | retina | AB1-FW18 | GEO | Homo sapiens | GSE86981 | Download (20,775) | |
| CTCF | retina | AB1-FW23 | GEO | Homo sapiens | GSE86981 | Download (23,702) | |
| CTCF | retina | AB1-RB | GEO | Homo sapiens | GSE86981 | Download (30,033) | |
| CTCF | MIA-PaCa-2 | GEO | Homo sapiens | GSE88734 | Download (30,770) | ||
| CTCF | HCT-116 | GEO | Homo sapiens | GSE92879 | Download (31,657) | ||
| CTCF | HAP1 | GEO | Homo sapiens | GSE94992 | Download (39,811) | ||
| CTCF | HAP1 | SCC4KO | GEO | Homo sapiens | GSE94992 | Download (30,908) | |
| CTCF | HAP1 | WaplKO-33 | GEO | Homo sapiens | GSE94992 | Download (43,210) | |
| CTCF | HAP1 | WaplKO-33_SCC4KO | GEO | Homo sapiens | GSE94992 | Download (44,152) | |
| CTCF | MCF-10A | GEO | Homo sapiens | GSE98551 | Download (56,648) | ||
| CTCF | THP-1 | macrophage_PMA | GEO | Homo sapiens | GSE96800 | Download (78,669) | |
| CTCF | THP-1 | monocytes | GEO | Homo sapiens | GSE96800 | Download (83,214) | |
| CTCF | HUES-64 | GEO | Homo sapiens | GSE97394 | Download (35,899) | ||
| CTCF | HUES-64 | DNMT-KO | GEO | Homo sapiens | GSE97394 | Download (42,396) | |
| CTCF | MCF-10AT1 | GEO | Homo sapiens | GSE98551 | Download (43,625) | ||
| CTCF | MCF-10CA1a | GEO | Homo sapiens | GSE98551 | Download (38,510) | ||
| CTCF | hESC | GEO | Homo sapiens | GSE20650 | Download (18,964) | ||
| CTCF | hESC | NAIVE | GEO | Homo sapiens | GSE69646 | Download (64,565) | |
| CTCF | hESC | PRIMED | GEO | Homo sapiens | GSE69646 | Download (29,644) | |
| CTCF | HSPC | GEO | Homo sapiens | GSE67783 | Download (317,836) | ||
| CTCF | islet | GEO | Homo sapiens | GSE23784 | Download (35,825) | ||
| CTCF | Jurkat | GEO | Homo sapiens | GSE68976 | Download (34,869) | ||
| CTCF | lymphocyte | blood | GEO | Homo sapiens | GSE46832 | Download (18,603) | |
| CTCF | macrophage | GEO | Homo sapiens | GSE118305 | Download (22,876) | ||
| CTCF | macrophage | ZIKVneg | GEO | Homo sapiens | GSE118305 | Download (6,787) | |
| CTCF | macrophage | ZIKVnpos | GEO | Homo sapiens | GSE118305 | Download (16,293) | |
| CTCF | monocyte | BLOOD | GEO | Homo sapiens | GSE43098 | Download (9,269) | |
| CTCF | monocyte | MACROPHAGE | GEO | Homo sapiens | GSE43098 | Download (1) | |
| CTCF | SUM159 | GEO | Homo sapiens | GSE46055 | Download (60,476) | ||
| CTCF | tlymphocyte | naive | GEO | Homo sapiens | GSE74850 | Download (323) | |
| CTCF | U2OS | GEO | Homo sapiens | GSE87831 | Download (2,989) | ||
| CTCF | VCaP | ETOH | GEO | Homo sapiens | GSE84432 | Download (43,549) | |
| CTCF | VCaP | R1881 | GEO | Homo sapiens | GSE84432 | Download (47,667) | |
| CTCF | BCBL-1 | GEO | Homo sapiens | GSE38411 | Download (81,229) | ||
| CTCF | 786-O | NORMOXIA | GEO | Homo sapiens | GSE78113 | Download (72,948) | |
| CTCF | delta-47 | GEO | Homo sapiens | GSE70764 | Download (35,391) | ||
| CTCF | GM04025 | B-lymphocytes | GEO | Homo sapiens | GSE111170 | Download (65,479) | |
| CTCF | GM09236 | _B-lymphocytes | GEO | Homo sapiens | GSE111170 | Download (66,595) | |
| CTCF | GM09237 | B-lymphocytes | GEO | Homo sapiens | GSE111170 | Download (71,346) | |
| CTCF | GM17942 | GEO | Homo sapiens | GSE76922 | Download (50,891) | ||
| CTCF | Hep-G2 | GEO | Homo sapiens | GSE111000 | Download (19,652) | ||
| CTCF | Hep-G2 | RELACS | GEO | Homo sapiens | GSE111000 | Download (9,584) | |
| CTCF | Hep-G2 | RELACS_10000cells | GEO | Homo sapiens | GSE111000 | Download (2,250) | |
| CTCF | ID00014 | GEO | Homo sapiens | GSE76922 | Download (53,629) | ||
| CTCF | ID00015 | GEO | Homo sapiens | GSE76922 | Download (31,042) | ||
| CTCF | ID00016 | GEO | Homo sapiens | GSE76922 | Download (50,493) | ||
| CTCF | IMR-5 | GEO | Homo sapiens | GSE78957 | Download (29,448) | ||
| CTCF | IMR-90 | GEO | Homo sapiens | GSE43070 | Download (37,599) | ||
| CTCF | K-562 | GEO | Homo sapiens | GSE110681 | Download (36,748) | ||
| CTCF | K-562 | GEO | Homo sapiens | GSE70482 | Download (51,763) | ||
| CTCF | K-562 | GEO | Homo sapiens | GSE70764 | Download (67,402) | ||
| CTCF | K-562 | HOXA11_dMQ1 | GEO | Homo sapiens | GSE90691 | Download (9,118) | |
| CTCF | K-562 | HOXA11_MQ1Q147L | GEO | Homo sapiens | GSE90691 | Download (3,434) | |
| CTCF | K-562 | HOXA13_dMQ1 | GEO | Homo sapiens | GSE90691 | Download (11,297) | |
| CTCF | K-562 | HOXA13_MQ1Q147L | GEO | Homo sapiens | GSE90691 | Download (3,982) | |
| CTCF | K-562 | RUNX1_dMQ1 | GEO | Homo sapiens | GSE90691 | Download (13,630) | |
| CTCF | K-562 | RUNX1_MQ1Q147L | GEO | Homo sapiens | GSE90691 | Download (647) | |
| CTCF | K-562 | GEO | Homo sapiens | GSE92879 | Download (29,283) | ||
| CTCF | K-562 | Dox | GEO | Homo sapiens | GSE92879 | Download (29,041) | |
| CTCF | LUHMES | GEO | Homo sapiens | GSE109706 | Download (107,501) | ||
| CTCF | MCF-7 | GEO | Homo sapiens | GSE70764 | Download (58,215) | ||
| CTCF | MCF-7 | HYPOXIA | GEO | Homo sapiens | GSE78113 | Download (49,573) | |
| CTCF | MCF-7 | NORMOXIA | GEO | Homo sapiens | GSE78113 | Download (52,371) | |
| CTCF | OVCAR-8 | GEO | Homo sapiens | GSE70764 | Download (71,435) | ||
| CTCF | RH4 | GEO | Homo sapiens | GSE83726 | Download (64,321) | ||
| CTCF | SK-MEL-147 | GEO | Homo sapiens | GSE94488 | Download (54,332) | ||
| CTCF | SK-N-SH | GEO | Homo sapiens | GSE76815 | Download (42,631) | ||
| CTCF | THP-1 | GEO | Homo sapiens | GSE69962 | Download (5,488) | ||
| CTCF | THP-1 | 2D3 | GEO | Homo sapiens | GSE69962 | Download (7,774) | |
| CTCF | WA01 | EMB | GEO | Homo sapiens | GSE52457 | Download (109,757) | |
| CTCF | WA01 | MESEN | GEO | Homo sapiens | GSE52457 | Download (101,811) | |
| CTCF | WA01 | MESEN_STEM | GEO | Homo sapiens | GSE52457 | Download (51,131) | |
| CTCF | WA01 | NEUR | GEO | Homo sapiens | GSE52457 | Download (60,007) | |
| CTCF | WA01 | TROP | GEO | Homo sapiens | GSE52457 | Download (93,301) | |
| CTCFL | NHDF | GEO | Homo sapiens | GSE70764 | Download (9) | ||
| CTCFL | delta-47 | GEO | Homo sapiens | GSE70764 | Download (15,015) | ||
| CTCFL | K-562 | GEO | Homo sapiens | GSE70764 | Download (31,986) | ||
| CTCFL | OVCAR-8 | GEO | Homo sapiens | GSE70764 | Download (34,447) | ||
| CTCFL | K-562 | ENCODE | Homo sapiens | ENCSR000BNK | Download (26,328) | ||
| CTNNB1 | LS180 | GEO | Homo sapiens | GSE31939 | Download (9,793) | ||
| CTNNB1 | LS180 | 125 | GEO | Homo sapiens | GSE31939 | Download (9,534) | |
| CTNNB1 | hESC | activinA_15h | GEO | Homo sapiens | GSE99202 | Download (600) | |
| CTNNB1 | hESC | YAP-_activinA_15h | GEO | Homo sapiens | GSE99202 | Download (1,060) | |
| CUX1 | GM12878 | ENCODE | Homo sapiens | ENCSR000DYR | Download (592) | ||
| CUX1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR049KIZ | Download (31,866) | ||
| CUX1 | K-562 | ENCODE | Homo sapiens | ENCSR000EFO | Download (4,553) | ||
| CUX1 | MCF-7 | ENCODE | Homo sapiens | ENCSR017CEO | Download (18,762) | ||
| CUX1 | K-562 | GEO | Homo sapiens | GSE92882 | Download (1,679) | ||
| CUX1 | K-562 | shCUX1-A | GEO | Homo sapiens | GSE92882 | Download (1,125) | |
| CUX1 | K-562 | shCUX1-B | GEO | Homo sapiens | GSE92882 | Download (596) | |
| CXXC4 | HEK293T | GEO | Homo sapiens | GSE42958 | Download (17,917) | ||
| DACH1 | K-562 | ENCODE | Homo sapiens | ENCSR030TJP | Download (13,947) | ||
| DAXX | PC-3 | GEO | Homo sapiens | GSE68647 | Download (57,551) | ||
| DAXX | PC-3 | DAXX | GEO | Homo sapiens | GSE68647 | Download (26,553) | |
| DDX20 | MCF-7 | ENCODE | Homo sapiens | ENCSR330ADN | Download (1,775) | ||
| DDX20 | K-562 | ENCODE | Homo sapiens | ENCSR382AIB | Download (3,607) | ||
| DDX20 | K-562 | ENCODE | Homo sapiens | ENCSR446LAV | Download (9,856) | ||
| DDX21 | HeLa | GEO | Homo sapiens | GSE89420 | Download (3,037) | ||
| DDX21 | HeLa | siTCOF1 | GEO | Homo sapiens | GSE89420 | Download (288) | |
| DDX5 | HeLa | GEO | Homo sapiens | GSE24126 | Download (1,175) | ||
| DDX5 | NTERA2 | GEO | Homo sapiens | GSE58641 | Download (52,788) | ||
| DEAF1 | K-562 | ENCODE | Homo sapiens | ENCSR387SYS | Download (2,340) | ||
| DEK | HeLa-S3 | ENCODE | Homo sapiens | ENCSR219MKK | Download (56,470) | ||
| DET1 | Col-0 | Col-0_seedling | 5d-D | GEO | Arabidopsis thaliana | GSE112951 | Download (12,305) |
| DET1 | Col-0 | Col-0_seedling | 5d-L | GEO | Arabidopsis thaliana | GSE112951 | Download (12,782) |
| DET1 | Col-0 | Col-0_seedling | det1_5d-D | GEO | Arabidopsis thaliana | GSE112951 | Download (12,173) |
| DET1 | Col-0 | Col-0_seedling | det1_5d-L | GEO | Arabidopsis thaliana | GSE112951 | Download (12,165) |
| DIDO1 | K-562 | ENCODE | Homo sapiens | ENCSR167JBG | Download (12,202) | ||
| DMAP1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR670YPQ | Download (47,981) | ||
| DNMT1 | K-562 | ENCODE | Homo sapiens | ENCSR987PBI | Download (1,628) | ||
| DNMT3B | Hep-G2 | ENCODE | Homo sapiens | ENCSR156CWW | Download (9,509) | ||
| DNMT3B | Hep-G2 | ENCODE | Homo sapiens | ENCSR283OLA | Download (18,910) | ||
| DNMT3B | HUES-8 | GEO | Homo sapiens | GSE99346 | Download (55,591) | ||
| DNMT3B | HUES-8 | TripleKO-TET1-2-3 | GEO | Homo sapiens | GSE99346 | Download (68,917) | |
| DOF2-2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (780) |
| DOF2-4 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (540) |
| DOF4-7 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (194) |
| DPBF3 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,258) |
| DPF1 | K-562 | GEO | Homo sapiens | GSE97661 | Download (6,724) | ||
| DPF1 | MCF-7 | GEO | Homo sapiens | GSE97661 | Download (4,054) | ||
| DPF2 | GM12878 | ENCODE | Homo sapiens | ENCSR509FWH | Download (56,816) | ||
| DPF2 | K-562 | ENCODE | Homo sapiens | ENCSR219BXP | Download (42,973) | ||
| DPF2 | K-562 | ENCODE | Homo sapiens | ENCSR715CCR | Download (86,114) | ||
| DPF2 | MCF-7 | ENCODE | Homo sapiens | ENCSR234VCE | Download (34,113) | ||
| DPPA3 | HEK293T | GEO | Homo sapiens | GSE53809 | Download (2) | ||
| DRAP1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR765MKZ | Download (69,807) | ||
| DRAP1 | GM12878 | GEO | Homo sapiens | GSE97661 | Download (10,653) | ||
| DREB1A | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,378) |
| DREB1B | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,704) |
| DREB1C | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,101) |
| DREB1C | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (12,005) |
| DREB1E | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (6,043) |
| DREB1F | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (421) |
| DREB2A | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (22,690) |
| DREB2A | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (3,602) |
| DREB2D | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (193) |
| DREB2E | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (456) |
| DUX4 | 22Rv1 | GEO | Homo sapiens | GSE95515 | Download (210,730) | ||
| DUX4 | HEK293 | GEO | Homo sapiens | GSE75791 | Download (9,068) | ||
| DUX4 | WA01 | GEO | Homo sapiens | GSE94322 | Download (75,655) | ||
| E2F1 | mesenchymal | GEO | Homo sapiens | GSE77260 | Download (14,504) | ||
| E2F1 | HMEC-1 | GEO | Homo sapiens | GSE62425 | Download (13,767) | ||
| E2F1 | LNCaP-abl | GEO | Homo sapiens | GSE67809 | Download (5,767) | ||
| E2F1 | MM1-S | GEO | Homo sapiens | GSE80661 | Download (9,544) | ||
| E2F1 | U266 | GEO | Homo sapiens | GSE80661 | Download (1,651) | ||
| E2F1 | LNCaP | shCON | GEO | Homo sapiens | GSE94958 | Download (14,534) | |
| E2F1 | LNCaP | shRB | GEO | Homo sapiens | GSE94958 | Download (35,445) | |
| E2F1 | MDA-MB-231 | GEO | Homo sapiens | GSE95303 | Download (50,450) | ||
| E2F1 | U-87MG | GBM | GEO | Homo sapiens | GSE99171 | Download (36,930) | |
| E2F1 | K-562 | ENCODE | Homo sapiens | ENCSR563LLO | Download (4,674) | ||
| E2F1 | K-562 | ENCODE | Homo sapiens | ENCSR720HUL | Download (25,980) | ||
| E2F1 | MCF-7 | ENCODE | Homo sapiens | ENCSR000EWX | Download (31,051) | ||
| E2F1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EVJ | Download (3,355) | ||
| E2F1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EVM | Download (11,071) | ||
| E2F1 | HeLa | GEO | Homo sapiens | GSE22478 | Download (14,154) | ||
| E2F3 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (378) |
| E2F4 | retina | pigment | GEO | Homo sapiens | GSE60024 | Download (705) | |
| E2F4 | MCF-10A | ENCODE | Homo sapiens | ENCSR000DOR | Download (18,806) | ||
| E2F4 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EVL | Download (1,660) | ||
| E2F4 | lymphoblastoid | GEO | Homo sapiens | GSE21488 | Download (533) | ||
| E2F4 | MCF-7 | ICI | GEO | Homo sapiens | GSE41561 | Download (916) | |
| E2F4 | MCF-7 | TAM | GEO | Homo sapiens | GSE41561 | Download (3,872) | |
| E2F4 | GM12878 | ENCODE | Homo sapiens | ENCSR000DYY | Download (939) | ||
| E2F4 | K-562 | ENCODE | Homo sapiens | ENCSR000EWL | Download (36,373) | ||
| E2F5 | K-562 | ENCODE | Homo sapiens | ENCSR709DRM | Download (3,888) | ||
| E2F6 | WA01 | ENCODE | Homo sapiens | ENCSR000BSI | Download (60,443) | ||
| E2F6 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EVK | Download (5,136) | ||
| E2F6 | A-549 | ENCODE | Homo sapiens | ENCSR000BTC | Download (20,372) | ||
| E2F6 | K-562 | ENCODE | Homo sapiens | ENCSR000BLI | Download (44,313) | ||
| E2F6 | K-562 | ENCODE | Homo sapiens | ENCSR000EWJ | Download (44,864) | ||
| E2F7 | K-562 | ENCODE | Homo sapiens | ENCSR171CAY | Download (916) | ||
| E2F7 | IMR-90 | GEO | Homo sapiens | GSE40343 | Download (596) | ||
| E2F7 | IMR-90 | QUIES | GEO | Homo sapiens | GSE40343 | Download (4,350) | |
| E2F7 | IMR-90 | SENES_E2F7 | GEO | Homo sapiens | GSE40343 | Download (3,810) | |
| E2F7 | IMR-90 | SENES_SHCTR | GEO | Homo sapiens | GSE40343 | Download (16,774) | |
| E2F7 | IMR-90 | SENES_SHRB | GEO | Homo sapiens | GSE40343 | Download (4,349) | |
| E2F8 | K-562 | ENCODE | Homo sapiens | ENCSR953DVM | Download (22,024) | ||
| E2F8 | MCF-7 | ENCODE | Homo sapiens | ENCSR897ZXU | Download (436) | ||
| E2F8 | GM12878 | ENCODE | Homo sapiens | ENCSR793HVL | Download (29,441) | ||
| E2FD | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (704) |
| E4F1 | GM12878 | ENCODE | Homo sapiens | ENCSR439WAF | Download (19,701) | ||
| E4F1 | K-562 | ENCODE | Homo sapiens | ENCSR731LHZ | Download (42,344) | ||
| E4F1 | MCF-7 | ENCODE | Homo sapiens | ENCSR841YWU | Download (4,842) | ||
| EBF1 | ASC | GEO | Homo sapiens | GSE54889 | Download (13,826) | ||
| EBF1 | LCL | GEO | Homo sapiens | GSE75503 | Download (20,604) | ||
| EBF1 | MUTUL | GEO | Homo sapiens | GSE75503 | Download (66,848) | ||
| EBF1 | GM12878 | ENCODE | Homo sapiens | ENCSR000BGU | Download (93,279) | ||
| EBF1 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZQ | Download (20,973) | ||
| EBF3 | SK-N-SH | GEO | Homo sapiens | GSE90682 | Download (36,390) | ||
| EBF3 | SK-N-SH | GEO | Homo sapiens | GSE90682 | Download (104,995) | ||
| EBS | Col-0 | Col-0_seedling | 10d | GEO | Arabidopsis thaliana | GSE104368 | Download (3,004) |
| EED | ProEs | GEO | Homo sapiens | GSE59087 | Download (69,377) | ||
| EED | GM12878 | ENCODE | Homo sapiens | ENCSR199WXF | Download (87,976) | ||
| EFM | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (4,979) |
| EGLN2 | T-47D | HAEGIN2 | GEO | Homo sapiens | GSE59935 | Download (95,380) | |
| EGR1 | A-375 | GEO | Homo sapiens | GSE116190 | Download (23,348) | ||
| EGR1 | GM12878 | ENCODE | Homo sapiens | ENCSR000BMQ | Download (23,084) | ||
| EGR1 | GM12878 | ENCODE | Homo sapiens | ENCSR000BRG | Download (2,128) | ||
| EGR1 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BSQ | Download (14,334) | ||
| EGR1 | liver | ENCODE | Homo sapiens | ENCSR290ZOS | Download (7,855) | ||
| EGR1 | liver | ENCODE | Homo sapiens | ENCSR736BUG | Download (20,152) | ||
| EGR1 | HL-60 | GEO | Homo sapiens | GSE106359 | Download (5,444) | ||
| EGR1 | HL-60 | PMA | GEO | Homo sapiens | GSE106359 | Download (7,062) | |
| EGR1 | K-562 | ENCODE | Homo sapiens | ENCSR000BNE | Download (45,782) | ||
| EGR1 | K-562 | ENCODE | Homo sapiens | ENCSR024CNP | Download (29,330) | ||
| EGR1 | K-562 | ENCODE | Homo sapiens | ENCSR211LTF | Download (29,625) | ||
| EGR1 | MCF-7 | ENCODE | Homo sapiens | ENCSR000BUX | Download (10,634) | ||
| EGR1 | WA01 | ENCODE | Homo sapiens | ENCSR000BJA | Download (9,407) | ||
| EGR1 | HCT-116 | ENCODE | Homo sapiens | ENCSR000BRZ | Download (10,812) | ||
| EGR2 | HEK293 | ENCODE | Homo sapiens | ENCSR919CZU | Download (80,949) | ||
| EHF | Calu-3 | GEO | Homo sapiens | GSE63398 | Download (151) | ||
| EHF | RWPE-1 | GEO | Homo sapiens | GSE114241 | Download (44,569) | ||
| EHF | primary-bronchial-epithelial | GEO | Homo sapiens | GSE85401 | Download (6,644) | ||
| EHMT2 | A-549 | ENCODE | Homo sapiens | ENCSR321BJQ | Download (2,983) | ||
| EHMT2 | K-562 | ENCODE | Homo sapiens | ENCSR175EOM | Download (33,384) | ||
| EICBP-B | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,482) |
| ELF1 | ME-1 | GEO | Homo sapiens | GSE46044 | Download (21,643) | ||
| ELF1 | GM12878 | ENCODE | Homo sapiens | ENCSR000BMB | Download (61,165) | ||
| ELF1 | GM12878 | ENCODE | Homo sapiens | ENCSR841NDX | Download (34,003) | ||
| ELF1 | A-549 | ENCODE | Homo sapiens | ENCSR000BPT | Download (17,370) | ||
| ELF1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BMZ | Download (40,349) | ||
| ELF1 | K-562 | ENCODE | Homo sapiens | ENCSR000BMD | Download (51,689) | ||
| ELF1 | K-562 | ENCODE | Homo sapiens | ENCSR502OEK | Download (13,262) | ||
| ELF1 | K-562 | ENCODE | Homo sapiens | ENCSR975SSR | Download (5,039) | ||
| ELF1 | MCF-7 | ENCODE | Homo sapiens | ENCSR000BSS | Download (31,378) | ||
| ELF1 | MCF-7 | ENCODE | Homo sapiens | ENCSR502NRF | Download (21,246) | ||
| ELF1 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BTA | Download (10,406) | ||
| ELF1 | HCT-116 | ENCODE | Homo sapiens | ENCSR000BVH | Download (4,357) | ||
| ELF1 | SK-N-MC | SHFLI_96H | GEO | Homo sapiens | GSE61944 | Download (40,166) | |
| ELF1 | SK-N-MC | SHGFP_96H | GEO | Homo sapiens | GSE61944 | Download (7,725) | |
| ELF3 | PDAC | GEO | Homo sapiens | GSE64557 | Download (66,738) | ||
| ELF3 | PDAC | KOKLF5 | GEO | Homo sapiens | GSE64557 | Download (52,323) | |
| ELF3 | PDAC | SHCTR | GEO | Homo sapiens | GSE64557 | Download (60,850) | |
| ELF3 | Hep-G2 | ENCODE | Homo sapiens | ENCSR770AOR | Download (104,537) | ||
| ELF4 | HEK293T | ENCODE | Homo sapiens | ENCSR778QLY | Download (3,614) | ||
| ELF4 | K-562 | ENCODE | Homo sapiens | ENCSR638QHV | Download (17,423) | ||
| ELF5 | T-47D | ELF5_48H | GEO | Homo sapiens | GSE31216 | Download (298) | |
| ELK1 | MCF-7 | ENCODE | Homo sapiens | ENCSR382WLL | Download (5,903) | ||
| ELK1 | WA01 | ENA | Homo sapiens | ERP002417 | Download (1,438) | ||
| ELK1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECI | Download (4,821) | ||
| ELK1 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZB | Download (914) | ||
| ELK1 | A-549 | ENCODE | Homo sapiens | ENCSR623KNM | Download (512) | ||
| ELK1 | IMR-90 | ENCODE | Homo sapiens | ENCSR664OKA | Download (1,285) | ||
| ELK1 | K-562 | ENCODE | Homo sapiens | ENCSR000EFU | Download (4,280) | ||
| ELK1 | K-562 | ENCODE | Homo sapiens | ENCSR338QAC | Download (5,518) | ||
| ELK4 | HEK293 | ENCODE | Homo sapiens | ENCSR000EVB | Download (303) | ||
| ELK4 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EVI | Download (7,923) | ||
| ELL | HCT-116 | GEO | Homo sapiens | GSE47938 | Download (610) | ||
| ELL2 | HCT-116 | SERUM | GEO | Homo sapiens | GSE30267 | Download (856) | |
| ELL2 | HCT-116 | STARVED | GEO | Homo sapiens | GSE30267 | Download (439) | |
| ELL2 | HeLa | GEO | Homo sapiens | GSE40632 | Download (38,060) | ||
| ELL2 | HeLa | DOX | GEO | Homo sapiens | GSE40632 | Download (43,350) | |
| ELL2 | HeLa | DOX_EGF | GEO | Homo sapiens | GSE40632 | Download (33,386) | |
| ELL2 | HeLa | EGF | GEO | Homo sapiens | GSE40632 | Download (19,390) | |
| EMB1789 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (227) |
| ENY | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (113) |
| EOMES | hESC | GEO | Homo sapiens | GSE26097 | Download (41,346) | ||
| EP300 | hESC | GEO | Homo sapiens | GSE17917 | Download (2,836) | ||
| EP300 | MCF-7 | DMSO | GEO | Homo sapiens | GSE29073 | Download (2,463) | |
| EP300 | MCF-7 | E2 | GEO | Homo sapiens | GSE29073 | Download (2,291) | |
| EP300 | MCF-7 | ENCODE | Homo sapiens | ENCSR000BTR | Download (15,788) | ||
| EP300 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BUA | Download (42,819) | ||
| EP300 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000EHV | Download (42,843) | ||
| EP300 | T-47D | ENCODE | Homo sapiens | ENCSR000BLM | Download (26,656) | ||
| EP300 | WA01 | ENCODE | Homo sapiens | ENCSR000AUQ | Download (9,125) | ||
| EP300 | WA01 | ENCODE | Homo sapiens | ENCSR000BKK | Download (19,843) | ||
| EP300 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECV | Download (33,577) | ||
| EP300 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR680OFU | Download (46,662) | ||
| EP300 | GM12878 | ENCODE | Homo sapiens | ENCSR000BHB | Download (9,864) | ||
| EP300 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZD | Download (5,603) | ||
| EP300 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZG | Download (6,680) | ||
| EP300 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BUE | Download (37,031) | ||
| EP300 | neural | ENCODE | Homo sapiens | ENCSR843ZUP | Download (48,009) | ||
| EP300 | osteoblast | ENCODE | Homo sapiens | ENCSR000AUD | Download (20,358) | ||
| EP300 | A-549 | ENCODE | Homo sapiens | ENCSR000BPW | Download (34,851) | ||
| EP300 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BLW | Download (55,105) | ||
| EP300 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EDV | Download (18,081) | ||
| EP300 | K-562 | ENCODE | Homo sapiens | ENCSR000AQB | Download (184) | ||
| EP300 | K-562 | ENCODE | Homo sapiens | ENCSR000EGE | Download (41,233) | ||
| EP300 | K-562 | ENCODE | Homo sapiens | ENCSR000EGY | Download (3,222) | ||
| EP400 | K-562 | ENCODE | Homo sapiens | ENCSR817QKV | Download (19,803) | ||
| EPAS1 | macrophage | HYPO_IL | GEO | Homo sapiens | GSE43109 | Download (1,145) | |
| EPAS1 | macrophage | NORMO_IL | GEO | Homo sapiens | GSE43109 | Download (325) | |
| EPAS1 | 786-O | GEO | Homo sapiens | GSE34871 | Download (1,150) | ||
| EPAS1 | HUVEC-C | GEO | Homo sapiens | GSE89836 | Download (109) | ||
| ERF | HAEC | TNFa_4h | GEO | Homo sapiens | GSE89970 | Download (7,984) | |
| ERF | VCaP | GEO | Homo sapiens | GSE98809 | Download (3,789) | ||
| ERF | VCaP | DOX | GEO | Homo sapiens | GSE98809 | Download (8,294) | |
| ERF003 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,002) |
| ERF010 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (821) |
| ERF011 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,398) |
| ERF012 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (133) |
| ERF014 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (376) |
| ERF015 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,124) |
| ERF017 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,879) |
| ERF019 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (684) |
| ERF021 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (5,398) |
| ERF023 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (227) |
| ERF027 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (4,011) |
| ERF027 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (9,154) |
| ERF035 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (6,643) |
| ERF037 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,734) |
| ERF038 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,928) |
| ERF039 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,855) |
| ERF043 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (905) |
| ERF043 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (999) |
| ERF086 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (134) |
| ERF086 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (661) |
| ERF087 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (426) |
| ERF087 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (5,632) |
| ERF091 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (509) |
| ERF095 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (115) |
| ERF109 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (540) |
| ERF11 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,507) |
| ERF115 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,361) |
| ERF115 | Ler | Ler_cell | suspension_2d | GEO | Arabidopsis thaliana | GSE48793 | Download (1,354) |
| ERF118 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,586) |
| ERF15 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,472) |
| ERF15 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (10,338) |
| ERF2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (644) |
| ERF3 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,518) |
| ERF4 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,183) |
| ERF5 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (476) |
| ERF8 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (765) |
| ERG | ME-1 | GEO | Homo sapiens | GSE46044 | Download (51,044) | ||
| ERG | HAEC | GEO | Homo sapiens | GSE89970 | Download (16,487) | ||
| ERG | VCaP | GEO | Homo sapiens | GSE98809 | Download (30,205) | ||
| ERG | Jurkat | GEO | Homo sapiens | GSE49091 | Download (64,045) | ||
| ERG | LNCaP | ERG | GEO | Homo sapiens | GSE110655 | Download (137,556) | |
| ERG | LNCaP | ERG-R367K | GEO | Homo sapiens | GSE110655 | Download (111,624) | |
| ERG | SKNO1 | GEO | Homo sapiens | GSE23730 | Download (71,274) | ||
| ERG | VCaP | shARID1A | GEO | Homo sapiens | GSE110655 | Download (56,545) | |
| ERG | VCaP | shCt | GEO | Homo sapiens | GSE110655 | Download (108,536) | |
| ERG | VCaP | shERG | GEO | Homo sapiens | GSE110655 | Download (33,534) | |
| ERG | VCaP | GEO | Homo sapiens | GSE28950 | Download (20,087) | ||
| ERG | VCaP | DHAT_18H | GEO | Homo sapiens | GSE28950 | Download (34,989) | |
| ERG | VCaP | DHAT_2H | GEO | Homo sapiens | GSE28950 | Download (24,295) | |
| ERG | VCaP | GEO | Homo sapiens | GSE49091 | Download (44,952) | ||
| ERG | VCaP | ETOH | GEO | Homo sapiens | GSE49091 | Download (63,013) | |
| ERG | VCaP | R1881 | GEO | Homo sapiens | GSE49091 | Download (62,411) | |
| ERG | VCaP | DHT | GEO | Homo sapiens | GSE79128 | Download (19,427) | |
| ERG | VCaP | SH1_DHT | GEO | Homo sapiens | GSE79128 | Download (16,679) | |
| ERG | VCaP | SH2_DHT | GEO | Homo sapiens | GSE79128 | Download (11,707) | |
| ERG | VCaP | SH3_DHT | GEO | Homo sapiens | GSE79128 | Download (11,526) | |
| ERG | HUVEC-C | GEO | Homo sapiens | GSE109625 | Download (846) | ||
| ERG | HUVEC-C | VEGF_12h | GEO | Homo sapiens | GSE109625 | Download (1,928) | |
| ERG | HUVEC-C | VEGF_1h | GEO | Homo sapiens | GSE109625 | Download (400) | |
| ERG | HUVEC-C | VEGF_4h | GEO | Homo sapiens | GSE109625 | Download (844) | |
| ERG | K-562 | GEO | Homo sapiens | GSE23730 | Download (14,650) | ||
| ERG | K-562 | DOX | GEO | Homo sapiens | GSE23730 | Download (26,207) | |
| ERG | MCF-7 | GEO | Homo sapiens | GSE23730 | Download (37,001) | ||
| ERG | RWPE-1 | GEO | Homo sapiens | GSE114241 | Download (34,189) | ||
| ERG | RWPE-1 | FLAG | GEO | Homo sapiens | GSE29808 | Download (1,047) | |
| ERG | RWPE-1 | GEO | Homo sapiens | GSE37752 | Download (7,547) | ||
| ERG | RWPE-1 | S96E | GEO | Homo sapiens | GSE86232 | Download (206) | |
| ERG | TSU-1621MT | GEO | Homo sapiens | GSE60477 | Download (45,704) | ||
| ERG | TSU-1621MT | ATRA | GEO | Homo sapiens | GSE60477 | Download (78,022) | |
| ERG | AMLPZ12 | GEO | Homo sapiens | GSE23730 | Download (31,875) | ||
| ERG | CD34 | NR29 | GEO | Homo sapiens | GSE23730 | Download (33,650) | |
| ERG2 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (95,585) | ||
| ERG3 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (136,710) | ||
| ESR1 | MSF-7 | GEO | Homo sapiens | GSE116768 | Download (148,359) | ||
| ESR1 | H3396 | E2 | GEO | Homo sapiens | GSE32349 | Download (3,775) | |
| ESR1 | H3396 | ETOH | GEO | Homo sapiens | GSE32349 | Download (1,375) | |
| ESR1 | MCF-7-Luc-Y537S | E2 | GEO | Homo sapiens | GSE78284 | Download (17,177) | |
| ESR1 | MCF-7-Luc-Y537S | EtOH | GEO | Homo sapiens | GSE78284 | Download (10,058) | |
| ESR1 | T-47D-A | E2 | GEO | Homo sapiens | GSE80358 | Download (856) | |
| ESR1 | T-47D-A | E2_R5020 | GEO | Homo sapiens | GSE80358 | Download (1,263) | |
| ESR1 | T-47D-B | GEO | Homo sapiens | GSE80358 | Download (3,038) | ||
| ESR1 | T-47D-B | E2 | GEO | Homo sapiens | GSE80358 | Download (5,482) | |
| ESR1 | T-47D-B | E2_R5020 | GEO | Homo sapiens | GSE80358 | Download (7,011) | |
| ESR1 | T-47D-B | R5020 | GEO | Homo sapiens | GSE80358 | Download (939) | |
| ESR1 | MCF7-Tet-On | shCtr9_E2 | GEO | Homo sapiens | GSE80728 | Download (1,273) | |
| ESR1 | pleural-effusion | GEO | Homo sapiens | GSE86538 | Download (24,764) | ||
| ESR1 | endometrioid-adenocarcinoma | tumor_1 | GEO | Homo sapiens | GSE94031 | Download (942) | |
| ESR1 | endometrioid-adenocarcinoma | tumor_2 | GEO | Homo sapiens | GSE94031 | Download (33,354) | |
| ESR1 | endometrioid-adenocarcinoma | tumor_3 | GEO | Homo sapiens | GSE94031 | Download (19,766) | |
| ESR1 | endometrioid-adenocarcinoma | tumor_4 | GEO | Homo sapiens | GSE94031 | Download (2,741) | |
| ESR1 | endometrioid-adenocarcinoma | tumor_5 | GEO | Homo sapiens | GSE94031 | Download (310) | |
| ESR1 | endometrioid-adenocarcinoma | tumor_6 | GEO | Homo sapiens | GSE94031 | Download (4,549) | |
| ESR1 | endometrioid-adenocarcinoma | tumor_F | GEO | Homo sapiens | GSE94031 | Download (12,811) | |
| ESR1 | endometrioid-adenocarcinoma | tumor_G | GEO | Homo sapiens | GSE94031 | Download (625) | |
| ESR1 | endometrioid-adenocarcinoma | tumor_I | GEO | Homo sapiens | GSE94031 | Download (608) | |
| ESR1 | endometrioid-adenocarcinoma | tumor_J | GEO | Homo sapiens | GSE94031 | Download (1,761) | |
| ESR1 | Ishikawa | GEO | Homo sapiens | GSE99905 | Download (35,757) | ||
| ESR1 | Ishikawa | EnhiE2 | GEO | Homo sapiens | GSE99905 | Download (15,873) | |
| ESR1 | MCF-7 | dox | GEO | Homo sapiens | GSE94493 | Download (16,415) | |
| ESR1 | MCF-7 | dox_E2 | GEO | Homo sapiens | GSE94493 | Download (33,979) | |
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE94493 | Download (10,930) | |
| ESR1 | MCF-7 | E2_talen | GEO | Homo sapiens | GSE94493 | Download (27,608) | |
| ESR1 | MCF-7 | talen | GEO | Homo sapiens | GSE94493 | Download (18,107) | |
| ESR1 | MCF-7 | dox | GEO | Homo sapiens | GSE94493 | Download (70,659) | |
| ESR1 | MCF-7 | dox_E2 | GEO | Homo sapiens | GSE94493 | Download (58,794) | |
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE94493 | Download (9,685) | |
| ESR1 | MCF-7 | E2_talen | GEO | Homo sapiens | GSE94493 | Download (27,722) | |
| ESR1 | MCF-7 | talen | GEO | Homo sapiens | GSE94493 | Download (27,797) | |
| ESR1 | MCF-7 | dox | GEO | Homo sapiens | GSE94493 | Download (49,123) | |
| ESR1 | MCF-7 | dox_E2 | GEO | Homo sapiens | GSE94493 | Download (89,703) | |
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE94493 | Download (63,032) | |
| ESR1 | MCF-7 | aldosterone | GEO | Homo sapiens | GSE99626 | Download (40,129) | |
| ESR1 | MCF-7 | DHT | GEO | Homo sapiens | GSE99626 | Download (32,377) | |
| ESR1 | MCF-7 | estradiol_45min | GEO | Homo sapiens | GSE99626 | Download (8,699) | |
| ESR1 | MCF-7 | estradiol_45min_ChIP-and-reChIP | GEO | Homo sapiens | GSE99626 | Download (14,605) | |
| ESR1 | MCF-7 | estradiol_45min_H4 | GEO | Homo sapiens | GSE99626 | Download (8,608) | |
| ESR1 | MCF-7 | estradiol_4h | GEO | Homo sapiens | GSE99626 | Download (71,970) | |
| ESR1 | MCF-7 | estradiol-aldosterone_45min | GEO | Homo sapiens | GSE99626 | Download (9,809) | |
| ESR1 | MCF-7 | estradiol-aldosterone_4h | GEO | Homo sapiens | GSE99626 | Download (26,663) | |
| ESR1 | MCF-7 | estradiol-Dex_4h | GEO | Homo sapiens | GSE99626 | Download (53,542) | |
| ESR1 | MCF-7 | estradiol-Dex_75min | GEO | Homo sapiens | GSE99626 | Download (25,840) | |
| ESR1 | MCF-7 | estradiol-DHT_45min | GEO | Homo sapiens | GSE99626 | Download (32,328) | |
| ESR1 | MCF-7 | estradiol-DHT_4h | GEO | Homo sapiens | GSE99626 | Download (68,212) | |
| ESR1 | MCF-7 | estradiol-progesterone_4h | GEO | Homo sapiens | GSE99626 | Download (36,425) | |
| ESR1 | MCF-7 | estradiol-R5020_45min | GEO | Homo sapiens | GSE99626 | Download (4,256) | |
| ESR1 | MCF-7 | vehicle_45min_H2 | GEO | Homo sapiens | GSE99626 | Download (4,231) | |
| ESR1 | MCF-7 | vehicle_45min_I2 | GEO | Homo sapiens | GSE99626 | Download (2,505) | |
| ESR1 | MCF-7 | vehicle_4h | GEO | Homo sapiens | GSE99626 | Download (36,157) | |
| ESR1 | MCF-7 | vehicle_60min | GEO | Homo sapiens | GSE99626 | Download (2,883) | |
| ESR1 | MDA-MB-231 | GEO | Homo sapiens | GSE95121 | Download (256) | ||
| ESR1 | MDA-MB-231 | 45min | GEO | Homo sapiens | GSE95121 | Download (6,487) | |
| ESR1 | MDA-MB-231 | LQ | GEO | Homo sapiens | GSE95121 | Download (690) | |
| ESR1 | MDA-MB-231 | LQ_45min | GEO | Homo sapiens | GSE95121 | Download (33,892) | |
| ESR1 | T-47D | dox | GEO | Homo sapiens | GSE94493 | Download (20,820) | |
| ESR1 | T-47D | dox_E2 | GEO | Homo sapiens | GSE94493 | Download (19,290) | |
| ESR1 | T-47D | E2 | GEO | Homo sapiens | GSE94493 | Download (23,569) | |
| ESR1 | T-47D | CR3flp | GEO | Homo sapiens | GSE99479 | Download (21,605) | |
| ESR1 | T-47D | flp-ctrl | GEO | Homo sapiens | GSE99479 | Download (17,952) | |
| ESR1 | MCF-7 | ENA | Homo sapiens | ERP000209 | Download (21,649) | ||
| ESR1 | MCF-7 | E2 | ENA | Homo sapiens | ERP000209 | Download (65,359) | |
| ESR1 | MCF-7 | ENA | Homo sapiens | ERP000380 | Download (1,455) | ||
| ESR1 | MCF-7 | E2 | ENA | Homo sapiens | ERP000380 | Download (16,175) | |
| ESR1 | MCF-7 | E2_TAM | ENA | Homo sapiens | ERP000380 | Download (13,475) | |
| ESR1 | MCF-7 | SHCRT_E2 | ENA | Homo sapiens | ERP000380 | Download (39,941) | |
| ESR1 | MCF-7 | SHFOXA1_E2 | ENA | Homo sapiens | ERP000380 | Download (19,505) | |
| ESR1 | MCF-7 | TAM | ENA | Homo sapiens | ERP000380 | Download (8,208) | |
| ESR1 | MCF-7 | TAMR | ENA | Homo sapiens | ERP000380 | Download (3,050) | |
| ESR1 | MCF-7 | TAMR_TAM | ENA | Homo sapiens | ERP000380 | Download (3,245) | |
| ESR1 | MCF-7 | ENA | Homo sapiens | ERP000783 | Download (94,238) | ||
| ESR1 | MCF-7 | E2 | ENA | Homo sapiens | ERP000901 | Download (32,184) | |
| ESR1 | MCF-7 | ENA | Homo sapiens | ERP002305 | Download (62) | ||
| ESR1 | MCF-7 | GLYC | ENA | Homo sapiens | ERP002305 | Download (25,890) | |
| ESR1 | MCF-7 | MRNAHIST | ENA | Homo sapiens | ERP002305 | Download (2,732) | |
| ESR1 | MCF-7 | LY2 | ENA | Homo sapiens | ERP003844 | Download (93,047) | |
| ESR1 | MCF-7 | ESR1_wildtype | GEO | Homo sapiens | GSE100074 | Download (16,752) | |
| ESR1 | MCF-7 | ESR1_wildtype_LTED | GEO | Homo sapiens | GSE100074 | Download (6,601) | |
| ESR1 | MCF-7 | ESR1_mutant_LTED | GEO | Homo sapiens | GSE100074 | Download (10,725) | |
| ESR1 | MCF-7 | shKMT2C | GEO | Homo sapiens | GSE100328 | Download (17,174) | |
| ESR1 | MCF-7 | shKMT2C_R | GEO | Homo sapiens | GSE100328 | Download (22,288) | |
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE102410 | Download (34,329) | |
| ESR1 | MCF-7 | Tamoxifen | GEO | Homo sapiens | GSE102410 | Download (33,587) | |
| ESR1 | MCF-7 | vehicle | GEO | Homo sapiens | GSE102410 | Download (7,258) | |
| ESR1 | MCF-7 | Fulvestrant_HC11 | GEO | Homo sapiens | GSE102882 | Download (16,922) | |
| ESR1 | MCF-7 | HC11 | GEO | Homo sapiens | GSE102882 | Download (41,567) | |
| ESR1 | MCF-7 | GEO | Homo sapiens | GSE103023 | Download (49,589) | ||
| ESR1 | MCF-7 | AZD2014 | GEO | Homo sapiens | GSE103023 | Download (42,547) | |
| ESR1 | MCF-7 | RAD001 | GEO | Homo sapiens | GSE103023 | Download (57,010) | |
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE108883 | Download (21,334) | |
| ESR1 | MCF-7 | E2_30min | GEO | Homo sapiens | GSE108883 | Download (15,708) | |
| ESR1 | MCF-7 | E2_3h | GEO | Homo sapiens | GSE108883 | Download (7,116) | |
| ESR1 | MCF-7 | EtOH | GEO | Homo sapiens | GSE108883 | Download (948) | |
| ESR1 | MCF-7 | EtOH_30min | GEO | Homo sapiens | GSE108883 | Download (2,713) | |
| ESR1 | MCF-7 | EtOH_3h | GEO | Homo sapiens | GSE108883 | Download (4,030) | |
| ESR1 | MCF-7 | ICI | GEO | Homo sapiens | GSE108883 | Download (7,661) | |
| ESR1 | MCF-7 | ICI_30min | GEO | Homo sapiens | GSE108883 | Download (8,371) | |
| ESR1 | MCF-7 | ICI_3h | GEO | Homo sapiens | GSE108883 | Download (6,248) | |
| ESR1 | SUM44PE | ESR1_wildtype | GEO | Homo sapiens | GSE100074 | Download (36,448) | |
| ESR1 | SUM44PE | _ESR1_mutant_LTED | GEO | Homo sapiens | GSE100074 | Download (52,951) | |
| ESR1 | T-47D | ENCODE | Homo sapiens | ENCSR000BJS | Download (35,547) | ||
| ESR1 | T-47D | ENCODE | Homo sapiens | ENCSR000BKN | Download (35,100) | ||
| ESR1 | T-47D | ENCODE | Homo sapiens | ENCSR000BLL | Download (4,556) | ||
| ESR1 | T-47D | ENCODE | Homo sapiens | ENCSR000BQD | Download (17,342) | ||
| ESR1 | T-47D | E2_TAM | ENA | Homo sapiens | ERP000380 | Download (837) | |
| ESR1 | MDA-MB-134 | GEO | Homo sapiens | GSE109103 | Download (5,658) | ||
| ESR1 | MDA-MB-134 | E2 | GEO | Homo sapiens | GSE109103 | Download (50,641) | |
| ESR1 | MDA-MB-134 | FI | GEO | Homo sapiens | GSE109103 | Download (35,450) | |
| ESR1 | breast | tumor_BADOUTCOME | GEO | Homo sapiens | GSE40867 | Download (49,359) | |
| ESR1 | breast | tumor_GOODOUTCOME | GEO | Homo sapiens | GSE40867 | Download (3,444) | |
| ESR1 | breast | tumor-xenograft_1_E2_P4 | GEO | Homo sapiens | GSE93108 | Download (3,598) | |
| ESR1 | breast | tumor-xenograft_2_E2 | GEO | Homo sapiens | GSE93108 | Download (3,333) | |
| ESR1 | breast | tumor-xenograft_3_E2 | GEO | Homo sapiens | GSE93108 | Download (2,881) | |
| ESR1 | breast | tumor-xenograft_3_E2_P4 | GEO | Homo sapiens | GSE93108 | Download (3,841) | |
| ESR1 | HCC1428 | TLED | GEO | Homo sapiens | GSE27300 | Download (1,668) | |
| ESR1 | Ishikawa | GEO | Homo sapiens | GSE109891 | Download (1,889) | ||
| ESR1 | Ishikawa | Dex_E2 | GEO | Homo sapiens | GSE109891 | Download (14,966) | |
| ESR1 | Ishikawa | E2 | GEO | Homo sapiens | GSE109891 | Download (23,362) | |
| ESR1 | U2OS | GEO | Homo sapiens | GSE26110 | Download (935) | ||
| ESR1 | U2OS | E2 | GEO | Homo sapiens | GSE26110 | Download (62,362) | |
| ESR1 | VCaP | E2 | GEO | Homo sapiens | GSE43985 | Download (3,438) | |
| ESR1 | VCaP | E2_ERA | GEO | Homo sapiens | GSE43985 | Download (14,416) | |
| ESR1 | ZR751 | SICTR_E2 | GEO | Homo sapiens | GSE40129 | Download (768) | |
| ESR1 | ZR751 | SIGATA_E2 | GEO | Homo sapiens | GSE40129 | Download (1,630) | |
| ESR1 | ZR751 | GEO | Homo sapiens | GSE72249 | Download (19,829) | ||
| ESR1 | ZR751 | E2 | GEO | Homo sapiens | GSE72249 | Download (33,172) | |
| ESR1 | MCF-7 | E2_45min | GEO | Homo sapiens | GSE109820 | Download (25,555) | |
| ESR1 | MCF-7 | E2_90min | GEO | Homo sapiens | GSE109820 | Download (26,736) | |
| ESR1 | MCF-7 | 800 | GEO | Homo sapiens | GSE115607 | Download (16,625) | |
| ESR1 | MCF-7 | DMSO | GEO | Homo sapiens | GSE115607 | Download (9,180) | |
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE115607 | Download (19,873) | |
| ESR1 | MCF-7 | Fulv | GEO | Homo sapiens | GSE115607 | Download (22,435) | |
| ESR1 | MCF-7 | H3B-5942 | GEO | Homo sapiens | GSE115607 | Download (17,244) | |
| ESR1 | MCF-7 | H3B-6545 | GEO | Homo sapiens | GSE115607 | Download (18,792) | |
| ESR1 | MCF-7 | RAD1901 | GEO | Homo sapiens | GSE115607 | Download (17,541) | |
| ESR1 | MCF-7 | s5942 | GEO | Homo sapiens | GSE115607 | Download (16,243) | |
| ESR1 | MCF-7 | Sat-H3B-6545 | GEO | Homo sapiens | GSE115607 | Download (16,713) | |
| ESR1 | MCF-7 | Tamoxifen | GEO | Homo sapiens | GSE115607 | Download (13,927) | |
| ESR1 | MCF-7 | GEO | Homo sapiens | GSE119057 | Download (65,999) | ||
| ESR1 | MCF-7 | ER_t10 | GEO | Homo sapiens | GSE119057 | Download (65,901) | |
| ESR1 | MCF-7 | ER_t20 | GEO | Homo sapiens | GSE119057 | Download (64,733) | |
| ESR1 | MCF-7 | ER_t30 | GEO | Homo sapiens | GSE119057 | Download (67,350) | |
| ESR1 | MCF-7 | ER_t40 | GEO | Homo sapiens | GSE119057 | Download (60,874) | |
| ESR1 | MCF-7 | ER_t50 | GEO | Homo sapiens | GSE119057 | Download (68,528) | |
| ESR1 | MCF-7 | ER_t60 | GEO | Homo sapiens | GSE119057 | Download (58,729) | |
| ESR1 | MCF-7 | ER_t70 | GEO | Homo sapiens | GSE119057 | Download (61,400) | |
| ESR1 | MCF-7 | ER_t80 | GEO | Homo sapiens | GSE119057 | Download (63,250) | |
| ESR1 | MCF-7 | ER_t90 | GEO | Homo sapiens | GSE119057 | Download (60,610) | |
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE119702 | Download (9,364) | |
| ESR1 | MCF-7 | E2+4OHT | GEO | Homo sapiens | GSE119702 | Download (20,482) | |
| ESR1 | MCF-7 | E2+4OHT_SRC-3 | GEO | Homo sapiens | GSE119702 | Download (48,636) | |
| ESR1 | MCF-7 | E2_SRC-3 | GEO | Homo sapiens | GSE119702 | Download (8,145) | |
| ESR1 | MCF-7 | shFbxo_E2 | GEO | Homo sapiens | GSE119702 | Download (18,831) | |
| ESR1 | MCF-7 | shFbxo_E2_4OHT | GEO | Homo sapiens | GSE119702 | Download (18,839) | |
| ESR1 | MCF-7 | shFbxo_E2_4OHT_SRC-3 | GEO | Homo sapiens | GSE119702 | Download (9,059) | |
| ESR1 | MCF-7 | shFbxo_E2_SRC-3 | GEO | Homo sapiens | GSE119702 | Download (12,536) | |
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE14664 | Download (65,766) | |
| ESR1 | MCF-7 | TLED | GEO | Homo sapiens | GSE27300 | Download (2,535) | |
| ESR1 | MCF-7 | GEO | Homo sapiens | GSE35109 | Download (6,909) | ||
| ESR1 | MCF-7 | SICTR_E2 | GEO | Homo sapiens | GSE40129 | Download (57,570) | |
| ESR1 | MCF-7 | SIGATA_E2 | GEO | Homo sapiens | GSE40129 | Download (59,847) | |
| ESR1 | MCF-7 | GEO | Homo sapiens | GSE41561 | Download (58,584) | ||
| ESR1 | MCF-7 | GEO | Homo sapiens | GSE45822 | Download (32,084) | ||
| ESR1 | MCF-7 | GEO | Homo sapiens | GSE48930 | Download (71,199) | ||
| ESR1 | MCF-7 | SHESR1 | GEO | Homo sapiens | GSE53531 | Download (74) | |
| ESR1 | MCF-7 | LETR | GEO | Homo sapiens | GSE54592 | Download (326) | |
| ESR1 | MCF-7 | LETR_ANDRO | GEO | Homo sapiens | GSE54592 | Download (94) | |
| ESR1 | MCF-7 | LY2_ETOH | GEO | Homo sapiens | GSE54592 | Download (42,906) | |
| ESR1 | MCF-7 | GEO | Homo sapiens | GSE54855 | Download (68) | ||
| ESR1 | MCF-7 | E2_10M | GEO | Homo sapiens | GSE54855 | Download (5,075) | |
| ESR1 | MCF-7 | E2_120M | GEO | Homo sapiens | GSE54855 | Download (2,040) | |
| ESR1 | MCF-7 | E2_2M | GEO | Homo sapiens | GSE54855 | Download (1,175) | |
| ESR1 | MCF-7 | E2_40M | GEO | Homo sapiens | GSE54855 | Download (4,925) | |
| ESR1 | MCF-7 | E2_5M | GEO | Homo sapiens | GSE54855 | Download (1,779) | |
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE55921 | Download (4,542) | |
| ESR1 | MCF-7 | E2_JQ1 | GEO | Homo sapiens | GSE55921 | Download (3,092) | |
| ESR1 | MCF-7 | GEO | Homo sapiens | GSE59530 | Download (1,456) | ||
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE59530 | Download (26,315) | |
| ESR1 | MCF-7 | E2_TNF | GEO | Homo sapiens | GSE59530 | Download (21,641) | |
| ESR1 | MCF-7 | SHCTR_E2_TNF | GEO | Homo sapiens | GSE59530 | Download (36,407) | |
| ESR1 | MCF-7 | SHFOXA1_E2_TNF | GEO | Homo sapiens | GSE59530 | Download (11,482) | |
| ESR1 | MCF-7 | TNF | GEO | Homo sapiens | GSE59530 | Download (815) | |
| ESR1 | MCF-7 | GEO | Homo sapiens | GSE60270 | Download (6,713) | ||
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE60270 | Download (31,005) | |
| ESR1 | MCF-7 | SHCTR | GEO | Homo sapiens | GSE60270 | Download (2,943) | |
| ESR1 | MCF-7 | SHCTR_E2 | GEO | Homo sapiens | GSE60270 | Download (14,668) | |
| ESR1 | MCF-7 | SHFOXA1_E2 | GEO | Homo sapiens | GSE60270 | Download (1,661) | |
| ESR1 | MCF-7 | SHGATA3_E2 | GEO | Homo sapiens | GSE60270 | Download (3,094) | |
| ESR1 | MCF-7 | SHCTR | GEO | Homo sapiens | GSE60270 | Download (52) | |
| ESR1 | MCF-7 | SHCTR_E2 | GEO | Homo sapiens | GSE60270 | Download (1,305) | |
| ESR1 | MCF-7 | SHRARS | GEO | Homo sapiens | GSE60270 | Download (346) | |
| ESR1 | MCF-7 | SHRARS_E2 | GEO | Homo sapiens | GSE60270 | Download (93) | |
| ESR1 | MCF-7 | E2_45m | GEO | Homo sapiens | GSE67295 | Download (69,743) | |
| ESR1 | MCF-7 | E2-ICI | GEO | Homo sapiens | GSE67295 | Download (8,800) | |
| ESR1 | MCF-7 | IKK7 | GEO | Homo sapiens | GSE67295 | Download (6,510) | |
| ESR1 | MCF-7 | IL1b_45m | GEO | Homo sapiens | GSE67295 | Download (35,731) | |
| ESR1 | MCF-7 | IL1b-ICI | GEO | Homo sapiens | GSE67295 | Download (15,532) | |
| ESR1 | MCF-7 | IL1b_IKK7 | GEO | Homo sapiens | GSE67295 | Download (3,186) | |
| ESR1 | MCF-7 | TNFa_45m | GEO | Homo sapiens | GSE67295 | Download (27,334) | |
| ESR1 | MCF-7 | Veh | GEO | Homo sapiens | GSE67295 | Download (41,854) | |
| ESR1 | MCF-7 | GEO | Homo sapiens | GSE68355 | Download (44,258) | ||
| ESR1 | MCF-7 | PROG | GEO | Homo sapiens | GSE68355 | Download (77,900) | |
| ESR1 | MCF-7 | R5020 | GEO | Homo sapiens | GSE68355 | Download (68,007) | |
| ESR1 | MCF-7 | GEO | Homo sapiens | GSE68356 | Download (9,313) | ||
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE68356 | Download (13,445) | |
| ESR1 | MCF-7 | E2PG | GEO | Homo sapiens | GSE68356 | Download (16,737) | |
| ESR1 | MCF-7 | GEO | Homo sapiens | GSE71276 | Download (1,711) | ||
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE71276 | Download (19,303) | |
| ESR1 | MCF-7 | GEO | Homo sapiens | GSE72249 | Download (3,439) | ||
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE72249 | Download (16,874) | |
| ESR1 | MCF-7 | GEO | Homo sapiens | GSE76893 | Download (69,855) | ||
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE81510 | Download (9,192) | |
| ESR1 | MCF-7 | E2_Dex | GEO | Homo sapiens | GSE81510 | Download (11,871) | |
| ESR1 | MCF-7 | ICI | GEO | Homo sapiens | GSE81510 | Download (2,732) | |
| ESR1 | MCF-7 | ICI_Dex | GEO | Homo sapiens | GSE81510 | Download (7,889) | |
| ESR1 | MCF-7 | E2 | GEO | Homo sapiens | GSE86538 | Download (20,767) | |
| ESR1 | MCF-7 | LTED | GEO | Homo sapiens | GSE86538 | Download (9,409) | |
| ESR1 | MCF-7 | LTED_E2 | GEO | Homo sapiens | GSE86538 | Download (25,547) | |
| ESR1 | MCF-7 | RUNX2_DOX | GEO | Homo sapiens | GSE86538 | Download (66,470) | |
| ESR1 | MCF-7 | TAMR | GEO | Homo sapiens | GSE86538 | Download (9,021) | |
| ESR1 | MCF-7 | TAMR_E2 | GEO | Homo sapiens | GSE86538 | Download (11,315) | |
| ESR1 | MCF-7 | GEO | Homo sapiens | GSE94023 | Download (18,260) | ||
| ESR1 | MCF-7 | E2-10min-ERalpha | GEO | Homo sapiens | GSE94023 | Download (55,509) | |
| ESR1 | MCF-7 | E2-1280-min-ERalpha | GEO | Homo sapiens | GSE94023 | Download (26,886) | |
| ESR1 | MCF-7 | E2-160min-ERalpha | GEO | Homo sapiens | GSE94023 | Download (24,204) | |
| ESR1 | MCF-7 | E2-20min-ERalpha | GEO | Homo sapiens | GSE94023 | Download (59,373) | |
| ESR1 | MCF-7 | E2-320min-ERalpha | GEO | Homo sapiens | GSE94023 | Download (37,319) | |
| ESR1 | MCF-7 | E2-40min-ERalpha | GEO | Homo sapiens | GSE94023 | Download (63,185) | |
| ESR1 | MCF-7 | E2-5min-ERalpha | GEO | Homo sapiens | GSE94023 | Download (66,065) | |
| ESR1 | MCF-7 | E2-640min-ERalpha | GEO | Homo sapiens | GSE94023 | Download (8,160) | |
| ESR1 | MCF-7 | E2-80min-ERalpha | GEO | Homo sapiens | GSE94023 | Download (51,894) | |
| ESR1 | MCF-7-Luc | E2 | GEO | Homo sapiens | GSE78284 | Download (21,988) | |
| ESR1 | MCF-7-Luc | EtOH | GEO | Homo sapiens | GSE78284 | Download (3,475) | |
| ESR1 | MDA-MB-134-VI | ESTROGEN | GEO | Homo sapiens | GSE51022 | Download (37,830) | |
| ESR1 | MDA-MB-134-VI | ETOH | GEO | Homo sapiens | GSE51022 | Download (654) | |
| ESR1 | T-47D | GEO | Homo sapiens | GSE116170 | Download (2,854) | ||
| ESR1 | T-47D | GEO | Homo sapiens | GSE68355 | Download (7,820) | ||
| ESR1 | T-47D | PROG | GEO | Homo sapiens | GSE68355 | Download (26,292) | |
| ESR1 | T-47D | R5020 | GEO | Homo sapiens | GSE68355 | Download (19,882) | |
| ESR1 | T-47D | E2 | GEO | Homo sapiens | GSE68356 | Download (1,859) | |
| ESR1 | T-47D | GEO | Homo sapiens | GSE72249 | Download (5,750) | ||
| ESR1 | T-47D | E2 | GEO | Homo sapiens | GSE72249 | Download (8,543) | |
| ESR1 | T-47D | GEO | Homo sapiens | GSE74033 | Download (10,622) | ||
| ESR1 | T-47D | GEO | Homo sapiens | GSE84593 | Download (7,360) | ||
| ESR1 | breast | glyc | ENA | Homo sapiens | ERP002305 | Download (210) | |
| ESR1 | breast | mrnahist | ENA | Homo sapiens | ERP002305 | Download (6,653) | |
| ESR1 | breast | tumor_Female_1 | GEO | Homo sapiens | GSE104399 | Download (6,891) | |
| ESR1 | breast | tumor_Female_2 | GEO | Homo sapiens | GSE104399 | Download (20,797) | |
| ESR1 | breast | tumor_Female_6 | GEO | Homo sapiens | GSE104399 | Download (16,048) | |
| ESR1 | breast | tumor_Female_8 | GEO | Homo sapiens | GSE104399 | Download (8,352) | |
| ESR1 | breast | tumor_Male_1 | GEO | Homo sapiens | GSE104399 | Download (11,294) | |
| ESR1 | breast | tumor_Male_10 | GEO | Homo sapiens | GSE104399 | Download (74,853) | |
| ESR1 | breast | tumor_Male_11 | GEO | Homo sapiens | GSE104399 | Download (31,287) | |
| ESR1 | breast | tumor_Male_12 | GEO | Homo sapiens | GSE104399 | Download (9,269) | |
| ESR1 | breast | tumor_Male_13 | GEO | Homo sapiens | GSE104399 | Download (10,030) | |
| ESR1 | breast | tumor_Male_14 | GEO | Homo sapiens | GSE104399 | Download (44,416) | |
| ESR1 | breast | tumor_Male_15 | GEO | Homo sapiens | GSE104399 | Download (21,805) | |
| ESR1 | breast | tumor_Male_16 | GEO | Homo sapiens | GSE104399 | Download (13,885) | |
| ESR1 | breast | tumor_Male_17 | GEO | Homo sapiens | GSE104399 | Download (4,487) | |
| ESR1 | breast | tumor_Male_18 | GEO | Homo sapiens | GSE104399 | Download (21,027) | |
| ESR1 | breast | tumor_Male_19 | GEO | Homo sapiens | GSE104399 | Download (37,516) | |
| ESR1 | breast | tumor_Male_20 | GEO | Homo sapiens | GSE104399 | Download (27,977) | |
| ESR1 | breast | tumor_Male_21 | GEO | Homo sapiens | GSE104399 | Download (50,289) | |
| ESR1 | breast | tumor_Male_22 | GEO | Homo sapiens | GSE104399 | Download (3,986) | |
| ESR1 | breast | tumor_Male_23 | GEO | Homo sapiens | GSE104399 | Download (3,741) | |
| ESR1 | breast | tumor_Male_24 | GEO | Homo sapiens | GSE104399 | Download (2,579) | |
| ESR1 | breast | tumor_Male_25 | GEO | Homo sapiens | GSE104399 | Download (2,833) | |
| ESR1 | breast | tumor_Male_26 | GEO | Homo sapiens | GSE104399 | Download (59,641) | |
| ESR1 | breast | tumor_Male_27 | GEO | Homo sapiens | GSE104399 | Download (1,018) | |
| ESR1 | breast | tumor_Male_28 | GEO | Homo sapiens | GSE104399 | Download (21,947) | |
| ESR1 | breast | tumor_Male_29 | GEO | Homo sapiens | GSE104399 | Download (4,607) | |
| ESR1 | breast | tumor_Male_3 | GEO | Homo sapiens | GSE104399 | Download (14,668) | |
| ESR1 | breast | tumor_Male_30 | GEO | Homo sapiens | GSE104399 | Download (13,600) | |
| ESR1 | breast | tumor_Male_4 | GEO | Homo sapiens | GSE104399 | Download (17,315) | |
| ESR1 | breast | tumor_Male_5 | GEO | Homo sapiens | GSE104399 | Download (19,951) | |
| ESR1 | breast | tumor_Male_6 | GEO | Homo sapiens | GSE104399 | Download (6,586) | |
| ESR1 | breast | tumor_Male_7 | GEO | Homo sapiens | GSE104399 | Download (27,069) | |
| ESR1 | breast | tumor_Male_8 | GEO | Homo sapiens | GSE104399 | Download (6,623) | |
| ESR1 | breast | tumor_Male_9 | GEO | Homo sapiens | GSE104399 | Download (9,764) | |
| ESR1 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BIY | Download (38,143) | ||
| ESR1 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BIZ | Download (3,719) | ||
| ESR1 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BKL | Download (36,198) | ||
| ESR1 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BQR | Download (39,473) | ||
| ESR1 | ZR751 | E2_TAM | ENA | Homo sapiens | ERP000380 | Download (7,643) | |
| ESR1 | ZR751 | ENA | Homo sapiens | ERP000783 | Download (40,613) | ||
| ESR2 | MDA-MB-231 | estradiol | GEO | Homo sapiens | GSE108979 | Download (30,205) | |
| ESR2 | MDA-MB-231 | LY500307 | GEO | Homo sapiens | GSE108979 | Download (8,703) | |
| ESR2 | MCF-7 | C412_E2 | GEO | Homo sapiens | GSE48096 | Download (1,801) | |
| ESRRA | SKBR3 | GEO | Homo sapiens | GSE81651 | Download (19,607) | ||
| ESRRA | SKBR3 | EGF | GEO | Homo sapiens | GSE81651 | Download (29,873) | |
| ESRRA | SKBR3 | HRG | GEO | Homo sapiens | GSE81651 | Download (42,092) | |
| ESRRA | BT-474 | GEO | Homo sapiens | GSE75876 | Download (3,755) | ||
| ESRRA | BT-474 | AICAR | GEO | Homo sapiens | GSE75876 | Download (14,158) | |
| ESRRA | BT-474 | GEO | Homo sapiens | GSE81651 | Download (43,600) | ||
| ESRRA | BT-474 | EGF | GEO | Homo sapiens | GSE81651 | Download (23,021) | |
| ESRRA | BT-474 | HRG | GEO | Homo sapiens | GSE81651 | Download (38,145) | |
| ESRRA | GM12878 | ENCODE | Homo sapiens | ENCSR000DYQ | Download (1,967) | ||
| ESRRA | A-549 | ENCODE | Homo sapiens | ENCSR473SUA | Download (4,813) | ||
| ESRRA | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEW | Download (1,563) | ||
| ESRRA | K-562 | ENCODE | Homo sapiens | ENCSR486IFJ | Download (28,798) | ||
| ESRRA | MCF-7 | ENCODE | Homo sapiens | ENCSR337NQP | Download (3,378) | ||
| ESRRA | MCF-7 | ENCODE | Homo sapiens | ENCSR954WVZ | Download (12,425) | ||
| ETS1 | OVCAR-8 | GEO | Homo sapiens | GSE101832 | Download (14,307) | ||
| ETS1 | HEY-A8 | GEO | Homo sapiens | GSE101832 | Download (13,671) | ||
| ETS1 | DU145 | GEO | Homo sapiens | GSE59021 | Download (9,069) | ||
| ETS1 | 786-O | GEO | Homo sapiens | GSE86092 | Download (32,141) | ||
| ETS1 | HUVEC-C | GEO | Homo sapiens | GSE109625 | Download (15,849) | ||
| ETS1 | HUVEC-C | VEGF_12h | GEO | Homo sapiens | GSE109625 | Download (39,675) | |
| ETS1 | HUVEC-C | VEGF_1h | GEO | Homo sapiens | GSE109625 | Download (16,135) | |
| ETS1 | HUVEC-C | VEGF_4h | GEO | Homo sapiens | GSE109625 | Download (32,400) | |
| ETS1 | HUVEC-C | GEO | Homo sapiens | GSE41166 | Download (25,378) | ||
| ETS1 | HUVEC-C | VEGF_12H | GEO | Homo sapiens | GSE41166 | Download (56,821) | |
| ETS1 | HUVEC-C | VEGF_1H | GEO | Homo sapiens | GSE41166 | Download (25,129) | |
| ETS1 | HUVEC-C | VEGF_4H | GEO | Homo sapiens | GSE41166 | Download (50,035) | |
| ETS1 | HUVEC-C | GEO | Homo sapiens | GSE93030 | Download (15,837) | ||
| ETS1 | HUVEC-C | 12h | GEO | Homo sapiens | GSE93030 | Download (39,641) | |
| ETS1 | HUVEC-C | 1h | GEO | Homo sapiens | GSE93030 | Download (16,124) | |
| ETS1 | HUVEC-C | 4h | GEO | Homo sapiens | GSE93030 | Download (32,395) | |
| ETS1 | HUVEC-C | modETS1 | GEO | Homo sapiens | GSE93030 | Download (3,045) | |
| ETS1 | PANC-1 | GEO | Homo sapiens | GSE59021 | Download (5,365) | ||
| ETS1 | RWPE-1 | GEO | Homo sapiens | GSE29808 | Download (335) | ||
| ETS1 | GM12878 | ENCODE | Homo sapiens | ENCSR000BKA | Download (9,524) | ||
| ETS1 | hESC | ENCODE | Homo sapiens | ENCSR534VHI | Download (13,523) | ||
| ETS1 | ALL-SIL | GEO | Homo sapiens | GSE102209 | Download (2,123) | ||
| ETS1 | A-549 | ENCODE | Homo sapiens | ENCSR000BPU | Download (18,564) | ||
| ETS1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR681WHQ | Download (22,000) | ||
| ETS1 | K-562 | ENCODE | Homo sapiens | ENCSR000BKQ | Download (28,436) | ||
| ETS2 | DU145 | ETS1KO | GEO | Homo sapiens | GSE86238 | Download (313) | |
| ETV1 | MDA-Pca-2b | GEO | Homo sapiens | GSE106624 | Download (33,696) | ||
| ETV1 | GIST | GEO | Homo sapiens | GSE22441 | Download (21,888) | ||
| ETV1 | GIST-T1 | GEO | Homo sapiens | GSE106624 | Download (42,115) | ||
| ETV1 | GIST48 | iFOXF1 | GEO | Homo sapiens | GSE106624 | Download (841) | |
| ETV1 | GIST48 | siSCR | GEO | Homo sapiens | GSE106624 | Download (3,378) | |
| ETV1 | K-562 | ENCODE | Homo sapiens | ENCSR277DMR | Download (12,255) | ||
| ETV1 | COLO-800 | GEO | Homo sapiens | GSE80443 | Download (26,951) | ||
| ETV1 | LNCaP | GEO | Homo sapiens | GSE47120 | Download (24,354) | ||
| ETV1 | A-375 | GEO | Homo sapiens | GSE80443 | Download (6,084) | ||
| ETV1 | GIST-T1 | GEO | Homo sapiens | GSE80443 | Download (14,557) | ||
| ETV1 | GIST882 | GEO | Homo sapiens | GSE80443 | Download (4,356) | ||
| ETV1 | RWPE-1 | FLAG | GEO | Homo sapiens | GSE29808 | Download (1,164) | |
| ETV4 | Hep-G2 | ENCODE | Homo sapiens | ENCSR714YZG | Download (27,624) | ||
| ETV5 | Hep-G2 | ENCODE | Homo sapiens | ENCSR096IIB | Download (109,726) | ||
| ETV6 | GM12878 | ENCODE | Homo sapiens | ENCSR597VGC | Download (6,117) | ||
| ETV6 | GM12878 | ENCODE | Homo sapiens | ENCSR626VUC | Download (26,696) | ||
| ETV6 | K-562 | ENCODE | Homo sapiens | ENCSR000FCE | Download (4,569) | ||
| ETV6 | K-562 | ENCODE | Homo sapiens | ENCSR124BJR | Download (6,720) | ||
| ETV6 | Reh | pCCL-ETV6-HA | GEO | Homo sapiens | GSE102785 | Download (7,329) | |
| ETV6 | GM12878 | GEO | Homo sapiens | GSE97661 | Download (29,845) | ||
| EVI1 | SKH1 | GEO | Homo sapiens | GSE87283 | Download (34,468) | ||
| EVI1 | SKH1 | 10d | GEO | Homo sapiens | GSE87283 | Download (24,164) | |
| EVI1 | SKH1 | RUNX1-EVI1_KD | GEO | Homo sapiens | GSE87283 | Download (5,661) | |
| EWSR1 | K-562 | ENCODE | Homo sapiens | ENCSR142YYA | Download (4,172) | ||
| EWSR1 | HEK293 | GEO | Homo sapiens | GSE73492 | Download (16) | ||
| EZA1 | Col-0 | Col-0_seedling | 10d | GEO | Arabidopsis thaliana | GSE108960 | Download (2,725) |
| EZH1 | ProEs | GEO | Homo sapiens | GSE59087 | Download (35,143) | ||
| EZH1 | ProEs | SHCTR | GEO | Homo sapiens | GSE59087 | Download (19,788) | |
| EZH1 | ProEs | SHEZH2 | GEO | Homo sapiens | GSE59087 | Download (6,119) | |
| EZH2 | hESC | GEO | Homo sapiens | GSE13084 | Download (119) | ||
| EZH2 | LNCaP | GEO | Homo sapiens | GSE39459 | Download (48,388) | ||
| EZH2 | Pfeiffer | GEO | Homo sapiens | GSE45982 | Download (8,687) | ||
| EZH2 | ProEs | GEO | Homo sapiens | GSE59087 | Download (8,048) | ||
| EZH2 | ProEs | SHCTR | GEO | Homo sapiens | GSE59087 | Download (7,584) | |
| EZH2 | ProEs | SHEZH2 | GEO | Homo sapiens | GSE59087 | Download (216) | |
| EZH2 | VCaP | DHAT_2H | GEO | Homo sapiens | GSE28950 | Download (3,682) | |
| EZH2 | VCaP | ETOH | GEO | Homo sapiens | GSE28950 | Download (2,758) | |
| EZH2 | HUVEC-C | GEO | Homo sapiens | GSE109625 | Download (7,208) | ||
| EZH2 | HUVEC-C | VEGF_12h | GEO | Homo sapiens | GSE109625 | Download (4,993) | |
| EZH2 | HUVEC-C | VEGF_1h | GEO | Homo sapiens | GSE109625 | Download (5,600) | |
| EZH2 | HUVEC-C | VEGF_4h | GEO | Homo sapiens | GSE109625 | Download (4,008) | |
| EZH2 | KG-1 | shPLZF | GEO | Homo sapiens | GSE109619 | Download (2,813) | |
| EZH2 | LNCaP-abl | GEO | Homo sapiens | GSE39459 | Download (51,451) | ||
| EZH2 | OCI-Ly1 | GEO | Homo sapiens | GSE45982 | Download (2,943) | ||
| EZH2 | OCI-Ly7 | GEO | Homo sapiens | GSE45982 | Download (1,341) | ||
| EZH2 | RH30 | DMSO | GEO | Homo sapiens | GSE85169 | Download (56,934) | |
| EZH2 | RH30 | Trametinib | GEO | Homo sapiens | GSE85169 | Download (360) | |
| EZH2 | SU-DHL-5 | GEO | Homo sapiens | GSE45982 | Download (68,655) | ||
| EZH2 | SU-DHL-6 | GEO | Homo sapiens | GSE45982 | Download (7,570) | ||
| EZH2 | T-REx-293 | K27WT | GEO | Homo sapiens | GSE118954 | Download (686) | |
| EZH2 | T-REx-293 | K27WT_12h | GEO | Homo sapiens | GSE118954 | Download (1,068) | |
| EZH2 | T-REx-293 | K27WT_24h | GEO | Homo sapiens | GSE118954 | Download (636) | |
| EZH2 | T-REx-293 | K27WT_6h | GEO | Homo sapiens | GSE118954 | Download (1,811) | |
| EZH2 | T-REx-293 | K27WT_72h | GEO | Homo sapiens | GSE118954 | Download (909) | |
| EZH2 | WSU-DLCL2 | GEO | Homo sapiens | GSE45982 | Download (22,730) | ||
| EZH2 | A-1847 | GEO | Homo sapiens | GSE95643 | Download (31,603) | ||
| EZH2 | A-1847 | CARM1-KO | GEO | Homo sapiens | GSE95643 | Download (74,674) | |
| EZH2 | K-562 | GEO | Homo sapiens | GSE97661 | Download (1,924) | ||
| EZH2 | SF8628 | GEO | Homo sapiens | GSE94834 | Download (22,498) | ||
| EZH2 | PC-9 | ENCODE | Homo sapiens | ENCSR793USK | Download (32,542) | ||
| EZH2 | SU-DHL-6 | ENCODE | Homo sapiens | ENCSR090TRI | Download (279) | ||
| EZH2 | WA01 | ENCODE | Homo sapiens | ENCSR000ASY | Download (15,914) | ||
| EZH2 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ATC | Download (134,327) | ||
| EZH2 | B-cell | ENCODE | Homo sapiens | ENCSR000AUZ | Download (5,031) | ||
| EZH2 | HCT-116 | ENCODE | Homo sapiens | ENCSR046HGP | Download (39,830) | ||
| EZH2 | farage | GEO | Homo sapiens | GSE45982 | Download (111,004) | ||
| EZH2 | astrocyte | ENCODE | Homo sapiens | ENCSR000ARR | Download (16,010) | ||
| EZH2 | DND41 | ENCODE | Homo sapiens | ENCSR000ASW | Download (47,509) | ||
| EZH2 | endothelial | umbilical-vein | ENCODE | Homo sapiens | ENCSR000ATA | Download (8,788) | |
| EZH2 | epithelial | mammary | ENCODE | Homo sapiens | ENCSR000ARE | Download (6,686) | |
| EZH2 | fibroblast | LUNG | ENCODE | Homo sapiens | ENCSR000ARO | Download (9,793) | |
| EZH2 | fibroblast | DERMAL | ENCODE | Homo sapiens | ENCSR000ASE | Download (7,543) | |
| EZH2 | GM12878 | ENCODE | Homo sapiens | ENCSR000ARD | Download (83) | ||
| EZH2 | hepatocyte | ENCODE | Homo sapiens | ENCSR384LYW | Download (7,682) | ||
| EZH2 | KARPAS422 | ENCODE | Homo sapiens | ENCSR646CKG | Download (153) | ||
| EZH2 | keratinocyte | ENCODE | Homo sapiens | ENCSR000ARK | Download (11,363) | ||
| EZH2 | myoblast | skeletal_muscle | ENCODE | Homo sapiens | ENCSR000ARH | Download (1,221) | |
| EZH2 | myotube | ENCODE | Homo sapiens | ENCSR000ASZ | Download (5,486) | ||
| EZH2 | neural | progenitor | ENCODE | Homo sapiens | ENCSR069DPL | Download (11,966) | |
| EZH2 | neural | ENCODE | Homo sapiens | ENCSR511CUH | Download (4,300) | ||
| EZH2 | A-673 | ENCODE | Homo sapiens | ENCSR179SAO | Download (13,549) | ||
| EZH2 | GM23248 | ENCODE | Homo sapiens | ENCSR131FFJ | Download (8,975) | ||
| EZH2 | GM23338 | ENCODE | Homo sapiens | ENCSR045YHA | Download (10,984) | ||
| EZH2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000ARI | Download (9,810) | ||
| EZH2 | K-562 | ENCODE | Homo sapiens | ENCSR000AQE | Download (1,797) | ||
| EZH2phosphoT487 | DOHH2 | ENCODE | Homo sapiens | ENCSR562NOP | Download (5,407) | ||
| EZH2phosphoT487 | hepatocyte | ENCODE | Homo sapiens | ENCSR341VYI | Download (20,883) | ||
| EZH2phosphoT487 | Loucy | ENCODE | Homo sapiens | ENCSR783QUL | Download (1,009) | ||
| EZH2phosphoT487 | neural | progenitor | ENCODE | Homo sapiens | ENCSR656MXA | Download (10,312) | |
| EZH2phosphoT487 | GM23248 | ENCODE | Homo sapiens | ENCSR096KPA | Download (16,547) | ||
| EZH2phosphoT487 | GM23338 | ENCODE | Homo sapiens | ENCSR591DTH | Download (1,215) | ||
| EZH2phosphoT487 | OCI-Ly3 | ENCODE | Homo sapiens | ENCSR584ATA | Download (268) | ||
| EZH2phosphoT487 | OCI-Ly7 | ENCODE | Homo sapiens | ENCSR565XSL | Download (8,118) | ||
| EZH2phosphoT487 | PC-3 | ENCODE | Homo sapiens | ENCSR617RSQ | Download (7,730) | ||
| EZH2phosphoT487 | PC-9 | ENCODE | Homo sapiens | ENCSR702GMW | Download (58,469) | ||
| EZH2phosphoT487 | SK-N-SH | ENCODE | Homo sapiens | ENCSR297MUV | Download (2,931) | ||
| EZH2phosphoT487 | SU-DHL-6 | ENCODE | Homo sapiens | ENCSR088HZI | Download (1,734) | ||
| EZH2phosphoT487 | HCT-116 | ENCODE | Homo sapiens | ENCSR429CLV | Download (6,928) | ||
| EZH2phosphoT487 | neuron | bipolar_doxy_4d | ENCODE | Homo sapiens | ENCSR886KKK | Download (24,095) | |
| FANCD2 | U2OS | APH_treated | GEO | Homo sapiens | GSE104464 | Download (1,892) | |
| FANCL | Jurkat | GEO | Homo sapiens | GSE45864 | Download (5,604) | ||
| FAR1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (705) |
| FEZF1 | HEK293 | ENCODE | Homo sapiens | ENCSR827NWO | Download (39,844) | ||
| FEZF1 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (41,439) | ||
| FHY3 | No-0 | No-0_seedling | 4d-dark | GEO | Arabidopsis thaliana | GSE30711 | Download (1,920) |
| FHY3 | No-0 | No-0_seedling | 4d-far-red | GEO | Arabidopsis thaliana | GSE30711 | Download (1,368) |
| FHY3 | Ler-0 | Ler-0_inflorescence | stage8-flower | GEO | Arabidopsis thaliana | GSE69422 | Download (640) |
| FIE | Ws | Ws_whole-plant | 30h-wt | GEO | Arabidopsis thaliana | GSE95562 | Download (2,643) |
| FIE | Col | Col_whole-plant | 30h | GEO | Arabidopsis thaliana | GSE84483 | Download (9,749) |
| FIP1L1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR313VZG | Download (37,804) | ||
| FIP1L1 | K-562 | ENCODE | Homo sapiens | ENCSR177DNR | Download (13,510) | ||
| FLC | Col-FRI | Col-FRI_aerial-part | 2w | GEO | Arabidopsis thaliana | GSE89889 | Download (936) |
| FLI1 | ME-1 | GEO | Homo sapiens | GSE46044 | Download (12,366) | ||
| FLI1 | A-673-clone-Asp114 | GEO | Homo sapiens | GSE92738 | Download (138,055) | ||
| FLI1 | A-673 | GEO | Homo sapiens | GSE99959 | Download (18,090) | ||
| FLI1 | NB4 | GEO | Homo sapiens | GSE23730 | Download (17,464) | ||
| FLI1 | SKNO1 | GEO | Homo sapiens | GSE23730 | Download (56,585) | ||
| FLI1 | UAE | GEO | Homo sapiens | GSE23730 | Download (27,398) | ||
| FLI1 | UAE | ZINC | GEO | Homo sapiens | GSE23730 | Download (31,833) | |
| FLI1 | A-673 | delta22_EWSFL-kd | GEO | Homo sapiens | GSE94480 | Download (125) | |
| FLI1 | A-673 | Mut9_EWSFL-kd | GEO | Homo sapiens | GSE94480 | Download (908) | |
| FLI1 | HUVEC-C | GEO | Homo sapiens | GSE109625 | Download (370) | ||
| FLI1 | HUVEC-C | VEGF_12h | GEO | Homo sapiens | GSE109625 | Download (5,750) | |
| FLI1 | HUVEC-C | VEGF_1h | GEO | Homo sapiens | GSE109625 | Download (4,289) | |
| FLI1 | HUVEC-C | VEGF_4h | GEO | Homo sapiens | GSE109625 | Download (1,174) | |
| FLI1 | SK-N-MC | GEO | Homo sapiens | GSE61944 | Download (3,020) | ||
| FLI1 | SK-N-MC | SHFLI_48H | GEO | Homo sapiens | GSE61944 | Download (717) | |
| FLI1 | SK-N-MC | SHFLI_96H | GEO | Homo sapiens | GSE61944 | Download (303) | |
| FLI1 | SK-N-MC | SHGFP_48H | GEO | Homo sapiens | GSE61944 | Download (4,299) | |
| FLI1 | SK-N-MC | SHGFP_96H | GEO | Homo sapiens | GSE61944 | Download (4,036) | |
| FLI1 | TSU-1621MT | GEO | Homo sapiens | GSE60477 | Download (28,326) | ||
| FLI1 | A-673 | 1_KRAB_eNKX2_1 | GEO | Homo sapiens | GSE106914 | Download (98,452) | |
| FLI1 | A-673 | 1_KRAB_GFP | GEO | Homo sapiens | GSE106914 | Download (71,478) | |
| FLI1 | A-673 | 2_KRAB_eSOX2_1 | GEO | Homo sapiens | GSE106914 | Download (52,563) | |
| FOS | endothelial | umbilical-vein | ENCODE | Homo sapiens | ENCSR000EVU | Download (60,061) | |
| FOS | GM12878 | ENCODE | Homo sapiens | ENCSR000EYZ | Download (1,970) | ||
| FOS | Hep-G2 | ENCODE | Homo sapiens | ENCSR177HDZ | Download (25,964) | ||
| FOS | IMR-90 | ENCODE | Homo sapiens | ENCSR124AIG | Download (59,276) | ||
| FOS | K-562 | ENCODE | Homo sapiens | ENCSR000DKB | Download (20,312) | ||
| FOS | K-562 | ENCODE | Homo sapiens | ENCSR000FAI | Download (14,761) | ||
| FOS | MG-63-3 | GEO | Homo sapiens | GSE74230 | Download (4,297) | ||
| FOS | MNNG-HOS | GEO | Homo sapiens | GSE74230 | Download (8,989) | ||
| FOS | MCF-10A | ENCODE | Homo sapiens | ENCSR000DON | Download (75,894) | ||
| FOS | MCF-10A | ENCODE | Homo sapiens | ENCSR000DOO | Download (83,604) | ||
| FOS | MCF-10A | ENCODE | Homo sapiens | ENCSR000DOP | Download (93,388) | ||
| FOS | MCF-10A | ENCODE | Homo sapiens | ENCSR000DOT | Download (82,161) | ||
| FOS | MCF-7 | ENCODE | Homo sapiens | ENCSR569XNP | Download (67,023) | ||
| FOS | THP-1 | eGFP-Pam3csk-0h | GEO | Homo sapiens | GSE103477 | Download (193) | |
| FOS | THP-1 | eGFP-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (6,061) | |
| FOS | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EZE | Download (32,273) | ||
| FOS | MCF10A-Er-Src | EtOH | GEO | Homo sapiens | GSE115597 | Download (104,017) | |
| FOS | MCF10A-Er-Src | TAM | GEO | Homo sapiens | GSE115597 | Download (138,820) | |
| FOS | MV4-11 | GEO | Homo sapiens | GSE64862 | Download (7,883) | ||
| FOS | MV4-11 | SHFLT3 | GEO | Homo sapiens | GSE64862 | Download (4,146) | |
| FOSL1 | BT-549 | GEO | Homo sapiens | GSE46166 | Download (25,448) | ||
| FOSL1 | 143B | GEO | Homo sapiens | GSE74230 | Download (16,859) | ||
| FOSL1 | MG-63-3 | GEO | Homo sapiens | GSE74230 | Download (9,954) | ||
| FOSL1 | MNNG-HOS | GEO | Homo sapiens | GSE74230 | Download (13,888) | ||
| FOSL1 | CD4-pos | resting | GEO | Homo sapiens | GSE87254 | Download (130) | |
| FOSL1 | CD4-pos | stimulated | GEO | Homo sapiens | GSE87254 | Download (109) | |
| FOSL1 | MDA-MB-231 | GEO | Homo sapiens | GSE95303 | Download (28,780) | ||
| FOSL1 | K-562 | ENCODE | Homo sapiens | ENCSR000BMV | Download (20,605) | ||
| FOSL1 | K-562 | ENCODE | Homo sapiens | ENCSR239ZLZ | Download (41,795) | ||
| FOSL1 | WA01 | ENCODE | Homo sapiens | ENCSR000BNS | Download (1,892) | ||
| FOSL1 | HCT-116 | ENCODE | Homo sapiens | ENCSR000BTE | Download (27,998) | ||
| FOSL2 | MCF-7 | ENCODE | Homo sapiens | ENCSR000BUI | Download (28,307) | ||
| FOSL2 | MCF-7 | ENCODE | Homo sapiens | ENCSR546KCN | Download (11,924) | ||
| FOSL2 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BVB | Download (56,044) | ||
| FOSL2 | hESC | GEO | Homo sapiens | GSE69539 | Download (42,662) | ||
| FOSL2 | HFOB | DIFF | GEO | Homo sapiens | GSE82295 | Download (2,106) | |
| FOSL2 | SK-MEL-147 | GEO | Homo sapiens | GSE94488 | Download (101,918) | ||
| FOSL2 | A-549 | ENCODE | Homo sapiens | ENCSR000BQO | Download (58,064) | ||
| FOSL2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BHP | Download (42,552) | ||
| FOXA1 | breast | tumor_Female_1 | GEO | Homo sapiens | GSE104399 | Download (22,681) | |
| FOXA1 | breast | tumor_Female_2 | GEO | Homo sapiens | GSE104399 | Download (34,634) | |
| FOXA1 | breast | tumor_Female_3 | GEO | Homo sapiens | GSE104399 | Download (13,654) | |
| FOXA1 | breast | tumor_Female_6 | GEO | Homo sapiens | GSE104399 | Download (3,658) | |
| FOXA1 | breast | tumor_Female_7 | GEO | Homo sapiens | GSE104399 | Download (6,415) | |
| FOXA1 | breast | tumor_Male_1 | GEO | Homo sapiens | GSE104399 | Download (16,031) | |
| FOXA1 | breast | tumor_Male_13 | GEO | Homo sapiens | GSE104399 | Download (27,246) | |
| FOXA1 | breast | tumor_Male_14 | GEO | Homo sapiens | GSE104399 | Download (49,170) | |
| FOXA1 | breast | tumor_Male_19 | GEO | Homo sapiens | GSE104399 | Download (31,975) | |
| FOXA1 | breast | tumor_Male_3 | GEO | Homo sapiens | GSE104399 | Download (19,078) | |
| FOXA1 | breast | tumor_Male_4 | GEO | Homo sapiens | GSE104399 | Download (26,909) | |
| FOXA1 | breast | tumor_Male_6 | GEO | Homo sapiens | GSE104399 | Download (6,949) | |
| FOXA1 | HEK293T | ENCODE | Homo sapiens | ENCSR094WHO | Download (518) | ||
| FOXA1 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BKW | Download (6,181) | ||
| FOXA1 | liver | ENCODE | Homo sapiens | ENCSR324RCI | Download (22,512) | ||
| FOXA1 | liver | ENCODE | Homo sapiens | ENCSR735KEY | Download (19,244) | ||
| FOXA1 | liver | ENA | Homo sapiens | ERP002306 | Download (49,212) | ||
| FOXA1 | LNCaP | ENA | Homo sapiens | ERP001226 | Download (106,487) | ||
| FOXA1 | ZR751 | E2_TAM | ENA | Homo sapiens | ERP000380 | Download (65,608) | |
| FOXA1 | A-549 | ENCODE | Homo sapiens | ENCSR000BPX | Download (63,580) | ||
| FOXA1 | A-549 | ENCODE | Homo sapiens | ENCSR000BRD | Download (39,541) | ||
| FOXA1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BLE | Download (102,115) | ||
| FOXA1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BMO | Download (85,953) | ||
| FOXA1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR267DFA | Download (7,834) | ||
| FOXA1 | K-562 | ENCODE | Homo sapiens | ENCSR819LHG | Download (332) | ||
| FOXA1 | MCF-7 | ENCODE | Homo sapiens | ENCSR126YEB | Download (49,194) | ||
| FOXA1 | MCF-7 | ENA | Homo sapiens | ERP000380 | Download (53,314) | ||
| FOXA1 | MCF-7 | E2_TAM | ENA | Homo sapiens | ERP000380 | Download (59,071) | |
| FOXA1 | MCF-7 | TAMR_E2_TAM | ENA | Homo sapiens | ERP000380 | Download (44,160) | |
| FOXA1 | MCF-7 | ENA | Homo sapiens | ERP001226 | Download (78,000) | ||
| FOXA1 | MDA-MB-453 | ENA | Homo sapiens | ERP001226 | Download (149,222) | ||
| FOXA1 | T-47D | ENCODE | Homo sapiens | ENCSR000BKY | Download (103,523) | ||
| FOXA1 | T-47D | E2_TAM | ENA | Homo sapiens | ERP000380 | Download (51,562) | |
| FOXA1 | 22Rv1 | EtOH | GEO | Homo sapiens | GSE80742 | Download (25,350) | |
| FOXA1 | 22Rv1 | R1881 | GEO | Homo sapiens | GSE80742 | Download (45,506) | |
| FOXA1 | 22Rv1 | GEO | Homo sapiens | GSE85558 | Download (23,687) | ||
| FOXA1 | 22Rv1 | Dox | GEO | Homo sapiens | GSE85558 | Download (48,174) | |
| FOXA1 | DU145 | GEO | Homo sapiens | GSE47987 | Download (4,223) | ||
| FOXA1 | DU145 | FOXA1 | GEO | Homo sapiens | GSE47987 | Download (105,720) | |
| FOXA1 | DU145 | FOXA1_ARC562S | GEO | Homo sapiens | GSE47987 | Download (119,448) | |
| FOXA1 | DU145 | FOXA1_ARQ6540X | GEO | Homo sapiens | GSE47987 | Download (93,843) | |
| FOXA1 | DU145 | FOXA1_HIGH | GEO | Homo sapiens | GSE47987 | Download (82,049) | |
| FOXA1 | A1A3 | Brg1KD_Dex | GEO | Homo sapiens | GSE112491 | Download (4,214) | |
| FOXA1 | A1A3 | Brg1KD_EtOH | GEO | Homo sapiens | GSE112491 | Download (3,007) | |
| FOXA1 | A1A3 | Dex | GEO | Homo sapiens | GSE112491 | Download (4,963) | |
| FOXA1 | A1A3 | EtOH | GEO | Homo sapiens | GSE112491 | Download (3,755) | |
| FOXA1 | T47D-A1-2 | Dex | GEO | Homo sapiens | GSE112491 | Download (6,740) | |
| FOXA1 | T47D-A1-2 | EtOH | GEO | Homo sapiens | GSE112491 | Download (2,779) | |
| FOXA1 | MCF-7-TAMR-1 | 4OHT | GEO | Homo sapiens | GSE113092 | Download (110,508) | |
| FOXA1 | MCF-7-TAMR-1 | E2 | GEO | Homo sapiens | GSE113092 | Download (159,501) | |
| FOXA1 | MCF-7-TAMR-1 | VEH | GEO | Homo sapiens | GSE113092 | Download (135,880) | |
| FOXA1 | MCF-7-WS8 | 4OHT | GEO | Homo sapiens | GSE113092 | Download (125,043) | |
| FOXA1 | MCF-7-WS8 | VEH | GEO | Homo sapiens | GSE113092 | Download (94,971) | |
| FOXA1 | LNCaP-C4-2B | GEO | Homo sapiens | GSE40050 | Download (12,297) | ||
| FOXA1 | LNCaP-C4-2B | DHT | GEO | Homo sapiens | GSE40050 | Download (19,883) | |
| FOXA1 | MCF-7-TAMR-1 | 4OH-Tam | GEO | Homo sapiens | GSE75201 | Download (66,798) | |
| FOXA1 | 22Rv1 | GEO | Homo sapiens | GSE96652 | Download (58,264) | ||
| FOXA1 | LNCaP | GEO | Homo sapiens | GSE94682 | Download (93,240) | ||
| FOXA1 | LNCaP | r1881 | GEO | Homo sapiens | GSE94682 | Download (98,660) | |
| FOXA1 | VCaP | GEO | Homo sapiens | GSE96652 | Download (293,244) | ||
| FOXA1 | VCaP | siERG | GEO | Homo sapiens | GSE96652 | Download (316,826) | |
| FOXA1 | T-47D | CR3flp | GEO | Homo sapiens | GSE99479 | Download (76,385) | |
| FOXA1 | T-47D | flp-ctrl | GEO | Homo sapiens | GSE99479 | Download (36,981) | |
| FOXA1 | LNCaP | DHT | GEO | Homo sapiens | GSE28264 | Download (48,486) | |
| FOXA1 | LNCaP | ETOH | GEO | Homo sapiens | GSE28264 | Download (82,844) | |
| FOXA1 | LNCaP | 1F5 | GEO | Homo sapiens | GSE30623 | Download (37,844) | |
| FOXA1 | LNCaP | 1F5_SIFOXA1 | GEO | Homo sapiens | GSE30623 | Download (9,934) | |
| FOXA1 | LNCaP | SHCTR_R1881 | GEO | Homo sapiens | GSE37345 | Download (100,698) | |
| FOXA1 | LNCaP | SHFOXA1_R1881 | GEO | Homo sapiens | GSE37345 | Download (79,528) | |
| FOXA1 | LNCaP | GEO | Homo sapiens | GSE52725 | Download (43,431) | ||
| FOXA1 | LNCaP | GEO | Homo sapiens | GSE56288 | Download (31,210) | ||
| FOXA1 | LNCaP | DHT24H | GEO | Homo sapiens | GSE58428 | Download (25,514) | |
| FOXA1 | LNCaP | ETOH24H | GEO | Homo sapiens | GSE58428 | Download (73,320) | |
| FOXA1 | LNCaP | GEO | Homo sapiens | GSE64656 | Download (78,192) | ||
| FOXA1 | LNCaP | ETOH | GEO | Homo sapiens | GSE69043 | Download (1,612) | |
| FOXA1 | LNCaP | PHFRPMIFCS | GEO | Homo sapiens | GSE69043 | Download (91,447) | |
| FOXA1 | LNCaP | R1881 | GEO | Homo sapiens | GSE69043 | Download (75,508) | |
| FOXA1 | LNCaP | SHGATA2_ETOH | GEO | Homo sapiens | GSE69043 | Download (146) | |
| FOXA1 | LNCaP | SHGATA2_R1881 | GEO | Homo sapiens | GSE69043 | Download (45,805) | |
| FOXA1 | LNCaP | DHT | GEO | Homo sapiens | GSE83860 | Download (201,192) | |
| FOXA1 | LNCaP | DHT_TNFA | GEO | Homo sapiens | GSE83860 | Download (248,728) | |
| FOXA1 | LNCaP | DMSO | GEO | Homo sapiens | GSE83860 | Download (194,305) | |
| FOXA1 | LNCaP | TNFA | GEO | Homo sapiens | GSE83860 | Download (245,271) | |
| FOXA1 | LNCaP | GEO | Homo sapiens | GSE85558 | Download (113,811) | ||
| FOXA1 | LNCaP | HNF4G_ovexp | GEO | Homo sapiens | GSE85558 | Download (118,239) | |
| FOXA1 | PDAC | GEO | Homo sapiens | GSE64557 | Download (49,994) | ||
| FOXA1 | PDAC | KOKLF5 | GEO | Homo sapiens | GSE64557 | Download (51,401) | |
| FOXA1 | PDAC | SHCTR | GEO | Homo sapiens | GSE64557 | Download (49,524) | |
| FOXA1 | prostate | tumor_tissue1 | GEO | Homo sapiens | GSE114385 | Download (1,186) | |
| FOXA1 | prostate | tumor_tissue2 | GEO | Homo sapiens | GSE114385 | Download (621) | |
| FOXA1 | prostate | tumor_tissue3 | GEO | Homo sapiens | GSE114385 | Download (2,113) | |
| FOXA1 | prostate | tumor_tissue4 | GEO | Homo sapiens | GSE114385 | Download (615) | |
| FOXA1 | VCaP | ETOH | GEO | Homo sapiens | GSE56086 | Download (234,228) | |
| FOXA1 | VCaP | R1881 | GEO | Homo sapiens | GSE56086 | Download (237,295) | |
| FOXA1 | VCaP | SHCTR_R1881 | GEO | Homo sapiens | GSE56086 | Download (198,856) | |
| FOXA1 | VCaP | SHPIAS1_R1881 | GEO | Homo sapiens | GSE56086 | Download (200,984) | |
| FOXA1 | VCaP | DHT24H | GEO | Homo sapiens | GSE58428 | Download (67,971) | |
| FOXA1 | VCaP | ETOH24H | GEO | Homo sapiens | GSE58428 | Download (48,984) | |
| FOXA1 | ZR751 | GEO | Homo sapiens | GSE72249 | Download (79,816) | ||
| FOXA1 | ZR751 | DEX | GEO | Homo sapiens | GSE72249 | Download (76,364) | |
| FOXA1 | ZR751 | E2 | GEO | Homo sapiens | GSE72249 | Download (67,789) | |
| FOXA1 | Huh-7 | ASYNC | GEO | Homo sapiens | GSE39241 | Download (5,115) | |
| FOXA1 | Huh-7 | MITO | GEO | Homo sapiens | GSE39241 | Download (60) | |
| FOXA1 | LNCaP-abl | GEO | Homo sapiens | GSE63034 | Download (71,936) | ||
| FOXA1 | LNCaP-abl | SHCREB1 | GEO | Homo sapiens | GSE63034 | Download (176,338) | |
| FOXA1 | LNCaP-abl | SHCTR | GEO | Homo sapiens | GSE63034 | Download (166,109) | |
| FOXA1 | MCF-7 | E2 | GEO | Homo sapiens | GSE23852 | Download (67,736) | |
| FOXA1 | MCF-7 | ETOH | GEO | Homo sapiens | GSE23852 | Download (60,158) | |
| FOXA1 | MCF-7 | SICTR | GEO | Homo sapiens | GSE40129 | Download (92,964) | |
| FOXA1 | MCF-7 | SIGATA | GEO | Homo sapiens | GSE40129 | Download (115,817) | |
| FOXA1 | MCF-7 | GEO | Homo sapiens | GSE59530 | Download (31,688) | ||
| FOXA1 | MCF-7 | E2 | GEO | Homo sapiens | GSE59530 | Download (33,650) | |
| FOXA1 | MCF-7 | E2_TNF | GEO | Homo sapiens | GSE59530 | Download (32,025) | |
| FOXA1 | MCF-7 | TNF | GEO | Homo sapiens | GSE59530 | Download (38,057) | |
| FOXA1 | MCF-7 | GEO | Homo sapiens | GSE60270 | Download (22,004) | ||
| FOXA1 | MCF-7 | E2 | GEO | Homo sapiens | GSE60270 | Download (77,723) | |
| FOXA1 | MCF-7 | GEO | Homo sapiens | GSE72249 | Download (65,783) | ||
| FOXA1 | MCF-7 | DEX | GEO | Homo sapiens | GSE72249 | Download (60,680) | |
| FOXA1 | MCF-7 | E2 | GEO | Homo sapiens | GSE72249 | Download (62,142) | |
| FOXA1 | MCF-7 | 4OH-Tam | GEO | Homo sapiens | GSE75201 | Download (55,123) | |
| FOXA1 | MCF-7 | GEO | Homo sapiens | GSE80808 | Download (39,207) | ||
| FOXA1 | MCF-7 | GEO | Homo sapiens | GSE81714 | Download (59,074) | ||
| FOXA1 | MCF-7 | siCT | GEO | Homo sapiens | GSE81714 | Download (155,777) | |
| FOXA1 | MDA-MB-453 | DHT | GEO | Homo sapiens | GSE45201 | Download (87,728) | |
| FOXA1 | NCI-H3122 | GEO | Homo sapiens | GSE39998 | Download (48,899) | ||
| FOXA1 | T-47D | GEO | Homo sapiens | GSE72249 | Download (26,680) | ||
| FOXA1 | T-47D | DEX | GEO | Homo sapiens | GSE72249 | Download (21,678) | |
| FOXA1 | T-47D | E2 | GEO | Homo sapiens | GSE72249 | Download (20,753) | |
| FOXA1 | T-47D | DMSO | GEO | Homo sapiens | GSE84593 | Download (63,567) | |
| FOXA2 | BJ1-hTERT | GEO | Homo sapiens | GSE90454 | Download (15,571) | ||
| FOXA2 | BJ1-hTERT | CDT1_Mimo | GEO | Homo sapiens | GSE90454 | Download (1,964) | |
| FOXA2 | BJ1-hTERT | FoxHnf1aCoExp | GEO | Homo sapiens | GSE90454 | Download (30,403) | |
| FOXA2 | BJ1-hTERT | GATA4 | GEO | Homo sapiens | GSE90454 | Download (6,890) | |
| FOXA2 | BJ1-hTERT | Mimo | GEO | Homo sapiens | GSE90454 | Download (21,416) | |
| FOXA2 | BJ1-hTERT | Mimo_Release | GEO | Homo sapiens | GSE90454 | Download (9,194) | |
| FOXA2 | BJ1-hTERT | MimosinePlus | GEO | Homo sapiens | GSE90454 | Download (26,241) | |
| FOXA2 | BJ1-hTERT | MimosineRelease | GEO | Homo sapiens | GSE90454 | Download (12,330) | |
| FOXA2 | BJ1-hTERT | Unind | GEO | Homo sapiens | GSE90454 | Download (4,255) | |
| FOXA2 | BJ1-hTERT | CDT1 | GEO | Homo sapiens | GSE92491 | Download (50,395) | |
| FOXA2 | BJ1-hTERT | FOXA2_GATA4_Coexp | GEO | Homo sapiens | GSE92491 | Download (79,179) | |
| FOXA2 | BJ1-hTERT | Mimo | GEO | Homo sapiens | GSE92491 | Download (52,049) | |
| FOXA2 | BJ1-hTERT | MimoRelease | GEO | Homo sapiens | GSE92491 | Download (44,756) | |
| FOXA2 | A-549 | GEO | Homo sapiens | GSE90454 | Download (101,277) | ||
| FOXA2 | Hep-G2 | GEO | Homo sapiens | GSE90454 | Download (83,889) | ||
| FOXA2 | CACO2 | GEO | Homo sapiens | GSE66218 | Download (49,051) | ||
| FOXA2 | islet | ENA | Homo sapiens | ERP004003 | Download (163,078) | ||
| FOXA2 | liver | ENCODE | Homo sapiens | ENCSR080XEY | Download (36,095) | ||
| FOXA2 | liver | ENCODE | Homo sapiens | ENCSR310NYI | Download (29,752) | ||
| FOXA2 | liver | CARN1618 | ENA | Homo sapiens | ERP008682 | Download (35,908) | |
| FOXA2 | pancreas | CARN1618 | ENA | Homo sapiens | ERP008682 | Download (6,954) | |
| FOXA2 | A-549 | ENCODE | Homo sapiens | ENCSR000BRE | Download (86,674) | ||
| FOXA2 | H9 | ENA | Homo sapiens | ERP004206 | Download (138,995) | ||
| FOXA2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BNI | Download (99,030) | ||
| FOXA2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR066EBK | Download (102,190) | ||
| FOXA2 | KerCT | GEO | Homo sapiens | GSE90454 | Download (71,142) | ||
| FOXA3 | Hep-G2 | ENCODE | Homo sapiens | ENCSR092OVN | Download (112,887) | ||
| FOXF1 | GIST-T1 | GEO | Homo sapiens | GSE106624 | Download (23,581) | ||
| FOXF1 | GIST48 | GEO | Homo sapiens | GSE106624 | Download (21,722) | ||
| FOXF1 | GIST48 | siETV1 | GEO | Homo sapiens | GSE106624 | Download (3,639) | |
| FOXJ2 | K-562 | ENCODE | Homo sapiens | ENCSR847LBF | Download (1,585) | ||
| FOXK1 | HEK293T | GEO | Homo sapiens | GSE51673 | Download (2,937) | ||
| FOXK2 | MCF-7 | ENCODE | Homo sapiens | ENCSR465VLK | Download (2,373) | ||
| FOXK2 | GM12878 | ENCODE | Homo sapiens | ENCSR861JUQ | Download (8,836) | ||
| FOXK2 | HEK293T | ENCODE | Homo sapiens | ENCSR872BOU | Download (210) | ||
| FOXK2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR171FUX | Download (12,799) | ||
| FOXK2 | K-562 | ENCODE | Homo sapiens | ENCSR302AWT | Download (23,510) | ||
| FOXK2 | K-562 | ENCODE | Homo sapiens | ENCSR508DQA | Download (25,442) | ||
| FOXM1 | GM12878 | ENCODE | Homo sapiens | ENCSR000BRU | Download (23,886) | ||
| FOXM1 | HEK293T | ENCODE | Homo sapiens | ENCSR831EIW | Download (1,498) | ||
| FOXM1 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BUS | Download (12,221) | ||
| FOXM1 | OE33 | ENA | Homo sapiens | ERP013564 | Download (2,058) | ||
| FOXM1 | K-562 | ENCODE | Homo sapiens | ENCSR429QPP | Download (17,679) | ||
| FOXM1 | MCF-7 | ENCODE | Homo sapiens | ENCSR000BUJ | Download (11,437) | ||
| FOXM1 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BTB | Download (8,379) | ||
| FOXM1 | HEK293 | GEO | Homo sapiens | GSE60032 | Download (2,625) | ||
| FOXM1 | HeLa | GEO | Homo sapiens | GSE52098 | Download (8,398) | ||
| FOXM1 | U2OS | GEO | Homo sapiens | GSE38170 | Download (342) | ||
| FOXM1 | HMEC-1 | GEO | Homo sapiens | GSE62425 | Download (112) | ||
| FOXM1 | MCF-7 | GEO | Homo sapiens | GSE40762 | Download (5,482) | ||
| FOXM1 | MCF-7 | THIOS | GEO | Homo sapiens | GSE40762 | Download (2,289) | |
| FOXM1 | MCF-7 | MI63 | GEO | Homo sapiens | GSE72977 | Download (896) | |
| FOXM1 | MDA-MB-231 | GEO | Homo sapiens | GSE40762 | Download (5,120) | ||
| FOXM1 | MDA-MB-231 | THIOS | GEO | Homo sapiens | GSE40762 | Download (12,812) | |
| FOXO1 | hESC | GEO | Homo sapiens | GSE69542 | Download (90,446) | ||
| FOXO1 | CD34 | GEO | Homo sapiens | GSE80773 | Download (56,256) | ||
| FOXO1 | B-cell | GERMINAL_CENTER | GEO | Homo sapiens | GSE68349 | Download (8,945) | |
| FOXO3 | Hep-G2 | GEO | Homo sapiens | GSE97661 | Download (22,065) | ||
| FOXO3 | K-562 | GEO | Homo sapiens | GSE97661 | Download (2,617) | ||
| FOXP1 | neural-progenitor | GEO | Homo sapiens | GSE62718 | Download (1,502) | ||
| FOXP1 | LNCaP | GEO | Homo sapiens | GSE62492 | Download (29,133) | ||
| FOXP1 | VCaP | DHT24H | GEO | Homo sapiens | GSE58428 | Download (88,596) | |
| FOXP1 | H9 | GEO | Homo sapiens | GSE31006 | Download (42,551) | ||
| FOXP1 | RCK8 | ENA | Homo sapiens | ERP010999 | Download (2) | ||
| FOXP1 | U2932 | ENA | Homo sapiens | ERP010999 | Download (8,399) | ||
| FOXP1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR029LBT | Download (9,322) | ||
| FOXP1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR369YUK | Download (60,107) | ||
| FOXP1 | SU-DHL-4 | ENA | Homo sapiens | ERP010999 | Download (345) | ||
| FOXP1 | SU-DHL-6 | ENA | Homo sapiens | ERP010999 | Download (15,515) | ||
| FOXP2 | SK-N-MC | ENCODE | Homo sapiens | ENCSR000BGB | Download (4) | ||
| FOXP2 | PFSK1 | ENCODE | Homo sapiens | ENCSR000BGA | Download (36,259) | ||
| FUS | Hep-G2 | ENCODE | Homo sapiens | ENCSR590QQP | Download (11,497) | ||
| FUS | K-562 | ENCODE | Homo sapiens | ENCSR051DXE | Download (4,930) | ||
| FUS3 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,168) |
| GABPA | THP-1 | calcitriol_100nM_1d | GEO | Homo sapiens | GSE98093 | Download (955) | |
| GABPA | THP-1 | EtOH_1d | GEO | Homo sapiens | GSE98093 | Download (1,231) | |
| GABPA | GM12878 | ENCODE | Homo sapiens | ENCSR000BGC | Download (4,565) | ||
| GABPA | GM12878 | ENCODE | Homo sapiens | ENCSR331HPA | Download (12,636) | ||
| GABPA | liver | ENCODE | Homo sapiens | ENCSR038GMB | Download (21,605) | ||
| GABPA | liver | ENCODE | Homo sapiens | ENCSR350ORK | Download (6,916) | ||
| GABPA | A-549 | ENCODE | Homo sapiens | ENCSR000BPY | Download (20,130) | ||
| GABPA | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BJK | Download (8,878) | ||
| GABPA | Hep-G2 | ENCODE | Homo sapiens | ENCSR269TNX | Download (16,192) | ||
| GABPA | HL-60 | ENCODE | Homo sapiens | ENCSR000BTK | Download (8,471) | ||
| GABPA | K-562 | ENCODE | Homo sapiens | ENCSR000BLO | Download (34,798) | ||
| GABPA | K-562 | ENCODE | Homo sapiens | ENCSR290MUH | Download (17,120) | ||
| GABPA | Jurkat | GEO | Homo sapiens | GSE49091 | Download (85,942) | ||
| GABPA | VCaP | GEO | Homo sapiens | GSE49091 | Download (54,951) | ||
| GABPA | VCaP | ETOH | GEO | Homo sapiens | GSE49091 | Download (52,957) | |
| GABPA | VCaP | R1881 | GEO | Homo sapiens | GSE49091 | Download (56,726) | |
| GABPA | Hep-G2 | GEO | Homo sapiens | GSE72082 | Download (16,631) | ||
| GABPA | MCF-7 | GEO | Homo sapiens | GSE72082 | Download (4,411) | ||
| GABPA | RWPE-1 | GEO | Homo sapiens | GSE29808 | Download (2,684) | ||
| GABPA | MCF-7 | ENCODE | Homo sapiens | ENCSR000BUK | Download (17,444) | ||
| GABPA | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BTG | Download (11,732) | ||
| GABPA | WA01 | ENCODE | Homo sapiens | ENCSR000BIW | Download (31,219) | ||
| GABPA | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000BHS | Download (9,892) | ||
| GABPA | DU145 | GEO | Homo sapiens | GSE59021 | Download (3,116) | ||
| GABPA | HeLa | GEO | Homo sapiens | GSE31417 | Download (2,957) | ||
| GABPB1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR462KYU | Download (87,040) | ||
| GABPB1 | K-562 | ENCODE | Homo sapiens | ENCSR138YYY | Download (32,437) | ||
| GATA1 | CD34 | Day7_30min | GEO | Homo sapiens | GSE104676 | Download (63,191) | |
| GATA1 | CD34 | Day9_30min | GEO | Homo sapiens | GSE104676 | Download (9,204) | |
| GATA1 | CHRF28811 | ENA | Homo sapiens | ERP008568 | Download (112) | ||
| GATA1 | erythroblast | ENCODE | Homo sapiens | ENCSR000EXP | Download (41,015) | ||
| GATA1 | HUDEP-2 | 30min | GEO | Homo sapiens | GSE104676 | Download (50,129) | |
| GATA1 | K-562 | ENCODE | Homo sapiens | ENCSR000EFT | Download (17,222) | ||
| GATA1 | K-562 | ENCODE | Homo sapiens | ENCSR000EWM | Download (10,873) | ||
| GATA1 | K-562 | GEO | Homo sapiens | GSE107726 | Download (66,089) | ||
| GATA1 | K-562 | MYO1D-Hub_KO | GEO | Homo sapiens | GSE107726 | Download (3,420) | |
| GATA1 | K-562 | MYO1D-Non-hub_KO | GEO | Homo sapiens | GSE107726 | Download (4,631) | |
| GATA1 | CD34 | ERYTH_BIO | GEO | Homo sapiens | GSE29194 | Download (18,763) | |
| GATA1 | CD34 | ERYTH_BMP | GEO | Homo sapiens | GSE29194 | Download (21,551) | |
| GATA1 | CD34 | GEO | Homo sapiens | GSE52924 | Download (193,372) | ||
| GATA1 | erythroid | R3R4 | GEO | Homo sapiens | GSE43625 | Download (10,416) | |
| GATA1 | HSPC-CD34pos | GEO | Homo sapiens | GSE93372 | Download (124,795) | ||
| GATA1 | K-562 | GEO | Homo sapiens | GSE70482 | Download (114,847) | ||
| GATA2 | LNCaP | GEO | Homo sapiens | GSE38391 | Download (79,591) | ||
| GATA2 | LNCaP | GEO | Homo sapiens | GSE52725 | Download (79,598) | ||
| GATA2 | LNCaP | CSFCS | GEO | Homo sapiens | GSE69043 | Download (72,939) | |
| GATA2 | LNCaP | ETOH | GEO | Homo sapiens | GSE69043 | Download (147,455) | |
| GATA2 | LNCaP | FBS | GEO | Homo sapiens | GSE69043 | Download (31,034) | |
| GATA2 | LNCaP | R1881 | GEO | Homo sapiens | GSE69043 | Download (147,708) | |
| GATA2 | LNCaP | SHGATA2_ETOH | GEO | Homo sapiens | GSE69043 | Download (118,033) | |
| GATA2 | LNCaP | SHGATA2_R1881 | GEO | Homo sapiens | GSE69043 | Download (134,920) | |
| GATA2 | TF1 | GEO | Homo sapiens | GSE73207 | Download (63,048) | ||
| GATA2 | HUVEC-C | GEO | Homo sapiens | GSE109625 | Download (4,827) | ||
| GATA2 | HUVEC-C | VEGF_12h | GEO | Homo sapiens | GSE109625 | Download (2,496) | |
| GATA2 | HUVEC-C | VEGF_1h | GEO | Homo sapiens | GSE109625 | Download (4,153) | |
| GATA2 | HUVEC-C | VEGF_4h | GEO | Homo sapiens | GSE109625 | Download (6,217) | |
| GATA2 | SKH1 | GEO | Homo sapiens | GSE87283 | Download (58,922) | ||
| GATA2 | SKH1 | RUNX1-EVI1_KD | GEO | Homo sapiens | GSE87283 | Download (37,229) | |
| GATA2 | TSU-1621MT | GEO | Homo sapiens | GSE60477 | Download (8,310) | ||
| GATA2 | SH-SY5Y | ENCODE | Homo sapiens | ENCSR000EYB | Download (52,017) | ||
| GATA2 | ESF | GEO | Homo sapiens | GSE108408 | Download (76,852) | ||
| GATA2 | CD34 | PROG_BMP | GEO | Homo sapiens | GSE29194 | Download (15) | |
| GATA2 | CD34 | ACY957 | GEO | Homo sapiens | GSE60792 | Download (34,619) | |
| GATA2 | CD34 | DMSO | GEO | Homo sapiens | GSE60792 | Download (8,092) | |
| GATA2 | endothelial | umbilical-vein | ENCODE | Homo sapiens | ENCSR000EVW | Download (34,627) | |
| GATA2 | WA09 | GEO | Homo sapiens | GSE105081 | Download (2,636) | ||
| GATA2 | HL-60 | ENA | Homo sapiens | ERP008568 | Download (51) | ||
| GATA2 | K-562 | ENCODE | Homo sapiens | ENCSR000BKM | Download (19,175) | ||
| GATA2 | K-562 | ENCODE | Homo sapiens | ENCSR000DKA | Download (36,100) | ||
| GATA2 | K-562 | ENCODE | Homo sapiens | ENCSR000EWG | Download (28,056) | ||
| GATA2 | ME-1 | GEO | Homo sapiens | GSE46044 | Download (12,947) | ||
| GATA20 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (21,121) |
| GATA21 | Col-0 | Col-0_whole-rosette-leaves | 3w | GEO | Arabidopsis thaliana | GSE97499 | Download (15,808) |
| GATA22 | Col-0 | Col-0_whole-rosette-leaves | 3w | GEO | Arabidopsis thaliana | GSE97499 | Download (1,702) |
| GATA3 | A1A3 | Brg1KD_Dex | GEO | Homo sapiens | GSE112491 | Download (5,606) | |
| GATA3 | A1A3 | Brg1KD_EtOH | GEO | Homo sapiens | GSE112491 | Download (5,866) | |
| GATA3 | A1A3 | Dex | GEO | Homo sapiens | GSE112491 | Download (4,175) | |
| GATA3 | A1A3 | EtOH | GEO | Homo sapiens | GSE112491 | Download (2,821) | |
| GATA3 | T47D-A1-2 | Dex | GEO | Homo sapiens | GSE112491 | Download (7,455) | |
| GATA3 | T47D-A1-2 | EtOH | GEO | Homo sapiens | GSE112491 | Download (7,404) | |
| GATA3 | Kelly | GEO | Homo sapiens | GSE94822 | Download (14,663) | ||
| GATA3 | T-47D | CR3flp | GEO | Homo sapiens | GSE99479 | Download (13,235) | |
| GATA3 | T-47D | flp-ctrl | GEO | Homo sapiens | GSE99479 | Download (6,993) | |
| GATA3 | T-47D | CR3flp | GEO | Homo sapiens | GSE99479 | Download (77,897) | |
| GATA3 | T-47D | flp-ctrl | GEO | Homo sapiens | GSE99479 | Download (28,022) | |
| GATA3 | SK-N-BE2-C | GEO | Homo sapiens | GSE94822 | Download (25,990) | ||
| GATA3 | MCF-7 | ENCODE | Homo sapiens | ENCSR000BST | Download (89,550) | ||
| GATA3 | MCF-7 | ENCODE | Homo sapiens | ENCSR000EWS | Download (16,224) | ||
| GATA3 | MCF-7 | ENCODE | Homo sapiens | ENCSR000EWV | Download (4,238) | ||
| GATA3 | MCF-7 | ENCODE | Homo sapiens | ENCSR423RTK | Download (81,256) | ||
| GATA3 | SH-SY5Y | ENCODE | Homo sapiens | ENCSR000EXZ | Download (24,426) | ||
| GATA3 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BTH | Download (121,502) | ||
| GATA3 | T-47D | ENCODE | Homo sapiens | ENCSR000BMX | Download (72,223) | ||
| GATA3 | BE2C | GEO | Homo sapiens | GSE65664 | Download (22,499) | ||
| GATA3 | CD4 | TH1 | GEO | Homo sapiens | GSE72266 | Download (191) | |
| GATA3 | CD4 | TH2 | GEO | Homo sapiens | GSE72266 | Download (5,368) | |
| GATA3 | Jurkat | GEO | Homo sapiens | GSE29180 | Download (5,230) | ||
| GATA3 | Jurkat | GEO | Homo sapiens | GSE68976 | Download (9,381) | ||
| GATA3 | Jurkat | GEO | Homo sapiens | GSE76181 | Download (49,867) | ||
| GATA3 | Kelly | GEO | Homo sapiens | GSE65664 | Download (27,058) | ||
| GATA3 | NGP | GEO | Homo sapiens | GSE65664 | Download (5,665) | ||
| GATA3 | RPMI8402 | GEO | Homo sapiens | GSE39179 | Download (440) | ||
| GATA3 | thymocyte | GEO | Homo sapiens | GSE71751 | Download (10,032) | ||
| GATA3 | CLB-Ga | GEO | Homo sapiens | GSE90683 | Download (16,545) | ||
| GATA3 | CCRF-CEM | GEO | Homo sapiens | GSE33850 | Download (870) | ||
| GATA3 | MCF-7 | DMSO | GEO | Homo sapiens | GSE29073 | Download (11,442) | |
| GATA3 | MCF-7 | E2 | GEO | Homo sapiens | GSE29073 | Download (23,765) | |
| GATA3 | MCF-7 | GEO | Homo sapiens | GSE40129 | Download (4,247) | ||
| GATA3 | MCF-7 | E2 | GEO | Homo sapiens | GSE40129 | Download (32,264) | |
| GATA3 | MCF-7 | GEO | Homo sapiens | GSE51274 | Download (47,989) | ||
| GATA3 | MCF-7 | GEO | Homo sapiens | GSE60270 | Download (903) | ||
| GATA3 | MCF-7 | E2 | GEO | Homo sapiens | GSE60270 | Download (14,121) | |
| GATA3 | MCF-7 | E2 | GEO | Homo sapiens | GSE81510 | Download (22,651) | |
| GATA3 | MCF-7 | E2_Dex | GEO | Homo sapiens | GSE81510 | Download (15,827) | |
| GATA3 | MCF-7 | ICI | GEO | Homo sapiens | GSE81510 | Download (4,615) | |
| GATA3 | SH-SY5Y | GEO | Homo sapiens | GSE65664 | Download (9,158) | ||
| GATA3 | T-47D | GEO | Homo sapiens | GSE51274 | Download (35,403) | ||
| GATA3 | breast | tumor_Male_15 | GEO | Homo sapiens | GSE104399 | Download (8,705) | |
| GATA3 | breast | tumor_Male_16 | GEO | Homo sapiens | GSE104399 | Download (6,674) | |
| GATA3 | breast | tumor_Male_21 | GEO | Homo sapiens | GSE104399 | Download (46,186) | |
| GATA3 | WA09 | GEO | Homo sapiens | GSE105081 | Download (365) | ||
| GATA3 | A-549 | ENCODE | Homo sapiens | ENCSR000BTI | Download (33,203) | ||
| GATA4 | Hep-G2 | ENCODE | Homo sapiens | ENCSR590CNM | Download (17,508) | ||
| GATA4 | AGS | GEO | Homo sapiens | GSE51705 | Download (92,924) | ||
| GATA4 | cardiomyocyte | GEO | Homo sapiens | GSE85628 | Download (4,973) | ||
| GATA4 | cardiomyocyte | 1 | GEO | Homo sapiens | GSE85628 | Download (5,259) | |
| GATA4 | cardiomyocyte | 5 | GEO | Homo sapiens | GSE85628 | Download (11,759) | |
| GATA4 | cardiomyocyte | 7 | GEO | Homo sapiens | GSE85628 | Download (9,981) | |
| GATA4 | G296S | GEO | Homo sapiens | GSE85628 | Download (11,034) | ||
| GATA4 | G296S | 2 | GEO | Homo sapiens | GSE85628 | Download (11,032) | |
| GATA4 | G296S | 4 | GEO | Homo sapiens | GSE85628 | Download (6,376) | |
| GATA4 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (773) |
| GATA4 | KATOIII | GEO | Homo sapiens | GSE51705 | Download (2,713) | ||
| GATA4 | BJ1-hTERT | GEO | Homo sapiens | GSE92491 | Download (72,424) | ||
| GATA4 | BJ1-hTERT | FOXA2_GATA4_Coexp | GEO | Homo sapiens | GSE92491 | Download (49,638) | |
| GATA4 | A-549 | GEO | Homo sapiens | GSE85002 | Download (9,869) | ||
| GATA4 | YCC-3 | GEO | Homo sapiens | GSE51705 | Download (6,962) | ||
| GATA6 | HUG1N | GEO | Homo sapiens | GSE51936 | Download (7,040) | ||
| GATA6 | KATOIII | GEO | Homo sapiens | GSE51705 | Download (2,518) | ||
| GATA6 | LS174T | GEO | Homo sapiens | GSE49320 | Download (3) | ||
| GATA6 | PATU8988 | GEO | Homo sapiens | GSE47535 | Download (38,407) | ||
| GATA6 | WA01 | GEO | Homo sapiens | GSE77360 | Download (81,100) | ||
| GATA6 | YCC-3 | GEO | Homo sapiens | GSE51705 | Download (33,437) | ||
| GATA6 | AGS | GEO | Homo sapiens | GSE51705 | Download (27,091) | ||
| GATA6 | AGS | GEO | Homo sapiens | GSE51936 | Download (35,027) | ||
| GATA6 | CACO2 | DIFF | GEO | Homo sapiens | GSE23436 | Download (480) | |
| GATA6 | CACO2 | PROLIF | GEO | Homo sapiens | GSE23436 | Download (14,834) | |
| GATA6 | H9 | ENA | Homo sapiens | ERP004206 | Download (2,027) | ||
| GATAD1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR108TYQ | Download (83,367) | ||
| GATAD1 | HeLa | GEO | Homo sapiens | GSE20303 | Download (8,779) | ||
| GATAD2A | Hep-G2 | ENCODE | Homo sapiens | ENCSR925BFV | Download (133,422) | ||
| GATAD2A | K-562 | ENCODE | Homo sapiens | ENCSR160QYK | Download (4,569) | ||
| GATAD2B | GM12878 | ENCODE | Homo sapiens | ENCSR828NCB | Download (38,611) | ||
| GATAD2B | K-562 | ENCODE | Homo sapiens | ENCSR547LKC | Download (5,777) | ||
| GATAD2B | MCF-7 | ENCODE | Homo sapiens | ENCSR389BLX | Download (465) | ||
| GATAD2B | MCF-7 | ENCODE | Homo sapiens | ENCSR979FQP | Download (21,924) | ||
| GBF2 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (19,778) |
| GBF2 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (19,998) |
| GBF3 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (20,095) |
| GBF3 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (21,728) |
| GBF3 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,014) |
| GBF3 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (4,682) |
| GFI1 | THP-1 | GEO | Homo sapiens | GSE90769 | Download (5,677) | ||
| GFI1 | THP-1 | OG86 | GEO | Homo sapiens | GSE90769 | Download (652) | |
| GFI1B | HEK293 | ENCODE | Homo sapiens | ENCSR445PDR | Download (8,700) | ||
| GFI1B | K-562 | ENCODE | Homo sapiens | ENCSR509GDT | Download (69,196) | ||
| GFI1B | CD34 | GEO | Homo sapiens | GSE52924 | Download (3,590) | ||
| GLI2 | HEK293 | ENCODE | Homo sapiens | ENCSR978EQY | Download (2,860) | ||
| GLI4 | HEK293 | ENCODE | Homo sapiens | ENCSR211PZO | Download (5,587) | ||
| GLI4 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (1,207) | ||
| GLIS1 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (100,475) | ||
| GLIS1 | HEK293 | ENCODE | Homo sapiens | ENCSR482BBZ | Download (79,873) | ||
| GLIS2 | HEK293 | ENCODE | Homo sapiens | ENCSR535DIA | Download (50,170) | ||
| GLYR1 | HeLa | GEO | Homo sapiens | GSE20303 | Download (20,814) | ||
| GMEB1 | K-562 | ENCODE | Homo sapiens | ENCSR928KOR | Download (23,891) | ||
| GMEB2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR745VSQ | Download (10,035) | ||
| GR | T47D-A1-2 | Dex | GEO | Homo sapiens | GSE112491 | Download (51,043) | |
| GRHL2 | MCF-7-TAMR-1 | VEH | GEO | Homo sapiens | GSE113092 | Download (83,014) | |
| GRHL2 | MCF-7-WS8 | VEH | GEO | Homo sapiens | GSE113092 | Download (52,683) | |
| GRHL2 | LNCaP | GEO | Homo sapiens | GSE80256 | Download (32,652) | ||
| GRHL2 | OVCA429 | GEO | Homo sapiens | GSE71018 | Download (36,518) | ||
| GRHL2 | PEO1 | GEO | Homo sapiens | GSE71018 | Download (22,773) | ||
| GRHL2 | MCF-7 | GEO | Homo sapiens | GSE109820 | Download (41,912) | ||
| GRHL2 | MCF-7 | E2_45min | GEO | Homo sapiens | GSE109820 | Download (42,970) | |
| GRHL2 | MCF-7 | GEO | Homo sapiens | GSE81714 | Download (83,442) | ||
| GRHL2 | OVCAR-3 | GEO | Homo sapiens | GSE71018 | Download (19,120) | ||
| GRHL2 | HBE | GEO | Homo sapiens | GSE46194 | Download (48,048) | ||
| GRHL3 | NHEKSV3 | GEO | Homo sapiens | GSE95199 | Download (493,031) | ||
| GSPT2 | HEK293T | GEO | Homo sapiens | GSE35197 | Download (19,952) | ||
| GT-1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (113) |
| GT-2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,643) |
| GT-2 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (4,943) |
| GT-3A | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (219) |
| GT-4 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (1,218) |
| GTF2A2 | K-562 | ENCODE | Homo sapiens | ENCSR801RPW | Download (2,250) | ||
| GTF2B | HeLa | ASYNC | GEO | Homo sapiens | GSE71848 | Download (19,350) | |
| GTF2B | HeLa | G2M | GEO | Homo sapiens | GSE71848 | Download (8,755) | |
| GTF2B | LCL | ENA | Homo sapiens | ERP004110 | Download (160,096) | ||
| GTF2B | K-562 | ENCODE | Homo sapiens | ENCSR000DOE | Download (2,023) | ||
| GTF2B | IMR-90 | TERT | GEO | Homo sapiens | GSE38303 | Download (15,011) | |
| GTF2F1 | MCF-7 | ENCODE | Homo sapiens | ENCSR557JTZ | Download (16,016) | ||
| GTF2F1 | WA01 | ENCODE | Homo sapiens | ENCSR000EBP | Download (6,942) | ||
| GTF2F1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECZ | Download (25,764) | ||
| GTF2F1 | GM12878 | ENCODE | Homo sapiens | ENCSR769ZTN | Download (17,338) | ||
| GTF2F1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR382PVA | Download (17,683) | ||
| GTF2F1 | K-562 | ENCODE | Homo sapiens | ENCSR000EHC | Download (7,293) | ||
| GTF2F1 | K-562 | ENCODE | Homo sapiens | ENCSR189VXS | Download (8,907) | ||
| GTF2F1 | K-562 | ENCODE | Homo sapiens | ENCSR377BLZ | Download (30,151) | ||
| GTF3A | HEK293 | GEO | Homo sapiens | GSE76494 | Download (113) | ||
| GTF3C2 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000DNY | Download (1,200) | ||
| GTF3C2 | K-562 | ENCODE | Homo sapiens | ENCSR000DOD | Download (7,039) | ||
| GTF3C5 | IMR-5 | CD532 | GEO | Homo sapiens | GSE78957 | Download (2,626) | |
| H1 | Col-0 | Col-0_seedling | 10d-H3-KD | GEO | Arabidopsis thaliana | GSE96834 | Download (27,326) |
| H2A | Col-0 | Col-0_leaves | 3w | GEO | Arabidopsis thaliana | GSE50942 | Download (13,163) |
| H2A-X | Col-0 | Col-0_leaves | 3w | GEO | Arabidopsis thaliana | GSE50942 | Download (9,098) |
| H2A-Z | Col-0 | Col-0_seedling | 6d-arp | GEO | Arabidopsis thaliana | GSE117391 | Download (268) |
| H2A-Z | Col-0 | Col-0_seedling | 6d-mbd | GEO | Arabidopsis thaliana | GSE117391 | Download (5,143) |
| H2A-Z | Col-0 | Col-0_seedling | 6d-wt | GEO | Arabidopsis thaliana | GSE117391 | Download (13,627) |
| H2A-Z | Col-0 | Col-0_seedling | 10d-H3-KD | GEO | Arabidopsis thaliana | GSE96834 | Download (29,817) |
| H2A-Z | Col-0 | Col-0_seedling | 10d-wt | GEO | Arabidopsis thaliana | GSE96834 | Download (28,897) |
| H2A-Z | Col-0 | Col-0_leaves | 3w | GEO | Arabidopsis thaliana | GSE50942 | Download (24,721) |
| H2A-Z | Col-0 | Col-0_leaves | GEO | Arabidopsis thaliana | GSE75071 | Download (26,406) | |
| H2A-Z | Col-0 | Col-0_aerial-part | 3w-clf-28 | GEO | Arabidopsis thaliana | GSE103361 | Download (25,384) |
| H2A-Z | Col-0 | Col-0_aerial-part | 3w-pie1-5 | GEO | Arabidopsis thaliana | GSE103361 | Download (22,990) |
| H2A-Z | Col-0 | Col-0_aerial-part | 3w-pkl-1 | GEO | Arabidopsis thaliana | GSE103361 | Download (26,848) |
| H2A-Z | Col-0 | Col-0_aerial-part | 3w-wt | GEO | Arabidopsis thaliana | GSE103361 | Download (27,240) |
| H2A-Z | Col-0 | Col-0_aerial-part | 12-13d-wt | GEO | Arabidopsis thaliana | GSE108450 | Download (22,378) |
| H2A-Z | Col-0 | Col-0_aerial-part | 13-16d-brm | GEO | Arabidopsis thaliana | GSE108450 | Download (20,935) |
| H2A-Z | Col-0 | Col-0_aerial-part | 12-14d-arp6 | GEO | Arabidopsis thaliana | GSE108450 | Download (9,139) |
| H2AK121ub | Col-0 | Col-0_seedling | 7d-atbmi | GEO | Arabidopsis thaliana | GSE89357 | Download (16,276) |
| H2AK121ub | Col-0 | Col-0_seedling | 7d-clf-swn | GEO | Arabidopsis thaliana | GSE89357 | Download (20,740) |
| H2AK121ub | Col-0 | Col-0_seedling | 7d-lhp1 | GEO | Arabidopsis thaliana | GSE89357 | Download (19,022) |
| H2AK121ub | Col-0 | Col-0_seedling | 7d-wt | GEO | Arabidopsis thaliana | GSE89357 | Download (19,632) |
| H2AV | Col-0 | Col-0_seedling | 12d-17c-15min | GEO | Arabidopsis thaliana | GSE79354 | Download (27,609) |
| H2AV | Col-0 | Col-0_seedling | 12d-17c-15min-exp | GEO | Arabidopsis thaliana | GSE79354 | Download (27,010) |
| H2AV | Col-0 | Col-0_seedling | 12d-17c-1h | GEO | Arabidopsis thaliana | GSE79354 | Download (27,093) |
| H2AV | Col-0 | Col-0_seedling | 12d-17c-1h-exp | GEO | Arabidopsis thaliana | GSE79354 | Download (27,284) |
| H2AV | Col-0 | Col-0_seedling | 12d-17c-4h | GEO | Arabidopsis thaliana | GSE79354 | Download (26,793) |
| H2AV | Col-0 | Col-0_seedling | 12d-17c-4h-exp | GEO | Arabidopsis thaliana | GSE79354 | Download (27,976) |
| H2AV | Col-0 | Col-0_seedling | 12d-27c-15min | GEO | Arabidopsis thaliana | GSE79354 | Download (27,131) |
| H2AV | Col-0 | Col-0_seedling | 12d-27c-1h | GEO | Arabidopsis thaliana | GSE79354 | Download (26,946) |
| H2AV | Col-0 | Col-0_seedling | 12d-27c-4h | GEO | Arabidopsis thaliana | GSE79354 | Download (27,791) |
| H2AV | Col-0 | Col-0_seedling | 12d-37c-15min-exp | GEO | Arabidopsis thaliana | GSE79354 | Download (27,574) |
| H2AV | Col-0 | Col-0_seedling | 12d-37c-1h-exp | GEO | Arabidopsis thaliana | GSE79354 | Download (27,622) |
| H2AV | Col-0 | Col-0_seedling | 12d-37c-4h-exp | GEO | Arabidopsis thaliana | GSE79354 | Download (27,295) |
| H2B | Col-0 | Col-0_seedling | 12d-17c-15min | GEO | Arabidopsis thaliana | GSE79354 | Download (31,555) |
| H2B | Col-0 | Col-0_seedling | 12d-17c-1h | GEO | Arabidopsis thaliana | GSE79354 | Download (28,146) |
| H2B | Col-0 | Col-0_seedling | 12d-17c-4h | GEO | Arabidopsis thaliana | GSE79354 | Download (31,344) |
| H2B | Col-0 | Col-0_seedling | 12d-27c-15min | GEO | Arabidopsis thaliana | GSE79354 | Download (34,006) |
| H2B | Col-0 | Col-0_seedling | 12d-27c-1h | GEO | Arabidopsis thaliana | GSE79354 | Download (18,416) |
| H2B | Col-0 | Col-0_seedling | 12d-27c-4h | GEO | Arabidopsis thaliana | GSE79354 | Download (29,848) |
| H3 | Col-0 | Col-0_seedling | 12d-17c-15min | GEO | Arabidopsis thaliana | GSE79354 | Download (13,471) |
| H3 | Col-0 | Col-0_seedling | 12d-17c-1h | GEO | Arabidopsis thaliana | GSE79354 | Download (16,102) |
| H3 | Col-0 | Col-0_seedling | 12d-17c-4h | GEO | Arabidopsis thaliana | GSE79354 | Download (19,524) |
| H3 | Col-0 | Col-0_seedling | 12d-27c-15min | GEO | Arabidopsis thaliana | GSE79354 | Download (19,174) |
| H3 | Col-0 | Col-0_seedling | 12d-27c-1h | GEO | Arabidopsis thaliana | GSE79354 | Download (21,768) |
| H3 | Col-0 | Col-0_seedling | 12d-27c-4h | GEO | Arabidopsis thaliana | GSE79354 | Download (15,593) |
| H3 | Col-0 | Col-0_seedling | 10-12d-kyp | GEO | Arabidopsis thaliana | GSE114009 | Download (18,600) |
| H3 | Col-0 | Col-0_seedling | 10-12d-kyp-suvh5-suvh6 | GEO | Arabidopsis thaliana | GSE114009 | Download (20,118) |
| H3 | Col-0 | Col-0_seedling | 10-12d-suvh5 | GEO | Arabidopsis thaliana | GSE114009 | Download (17,152) |
| H3 | Col-0 | Col-0_seedling | 10-12d-suvh56 | GEO | Arabidopsis thaliana | GSE114009 | Download (12,182) |
| H3 | Col-0 | Col-0_seedling | 10-12d-wt | GEO | Arabidopsis thaliana | GSE114009 | Download (19,517) |
| H3 | Col-0 | Col-0_seedling | 18d-hda19 | GEO | Arabidopsis thaliana | GSE124414 | Download (16,580) |
| H3 | Col-0 | Col-0_seedling | 18d-hda6 | GEO | Arabidopsis thaliana | GSE124414 | Download (19,500) |
| H3 | Col-0 | Col-0_seedling | 18d-hdc1 | GEO | Arabidopsis thaliana | GSE124414 | Download (17,116) |
| H3 | Col-0 | Col-0_seedling | 18d-wt | GEO | Arabidopsis thaliana | GSE124414 | Download (16,565) |
| H3 | Col-0 | Col-0_seedling | 3w-crh6 | GEO | Arabidopsis thaliana | GSE37644 | Download (30,440) |
| H3 | Col-0 | Col-0_seedling | 3w-crt1 | GEO | Arabidopsis thaliana | GSE37644 | Download (33,528) |
| H3 | Col-0 | Col-0_seedling | 3w-wt | GEO | Arabidopsis thaliana | GSE37644 | Download (32,220) |
| H3 | Col-0 | Col-0_seedling | 10d-H3-KD | GEO | Arabidopsis thaliana | GSE96834 | Download (32,022) |
| H3 | Col-0 | Col-0_endosperm | 4d-wt-x-wt | GEO | Arabidopsis thaliana | GSE100011 | Download (6,736) |
| H3 | Col-0 | Col-0_aerial-part | 3w-clf-28 | GEO | Arabidopsis thaliana | GSE103361 | Download (29,036) |
| H3 | Col-0 | Col-0_aerial-part | 3w-pie1-5 | GEO | Arabidopsis thaliana | GSE103361 | Download (24,595) |
| H3 | Col-0 | Col-0_aerial-part | 3w-pkl-1 | GEO | Arabidopsis thaliana | GSE103361 | Download (24,726) |
| H3 | Col-0 | Col-0_aerial-part | 3w-wt | GEO | Arabidopsis thaliana | GSE103361 | Download (21,459) |
| H3 | Col-0 | Col-0_aerial-part | 6d-caa39 | GEO | Arabidopsis thaliana | GSE103924 | Download (21,949) |
| H3 | Col-0 | Col-0_aerial-part | 6d-wt | GEO | Arabidopsis thaliana | GSE103924 | Download (17,151) |
| H3 | Col-0 | Col-0_rosette-leaves | 21d-mutant-drought | GEO | Arabidopsis thaliana | GSE68370 | Download (23,822) |
| H3 | Col-0 | Col-0_rosette-leaves | 21d-mutant-watered | GEO | Arabidopsis thaliana | GSE68370 | Download (23,260) |
| H3 | Col-0 | Col-0_rosette-leaves | 21d-wt-drought | GEO | Arabidopsis thaliana | GSE68370 | Download (33,789) |
| H3 | Col-0 | Col-0_rosette-leaves | 21d-wt-watered | GEO | Arabidopsis thaliana | GSE68370 | Download (33,501) |
| H3 | Col-x-Ler | Col-x-Ler_endosperm | 20d-1 | GEO | Arabidopsis thaliana | GSE66585 | Download (342) |
| H3 | Col-0 | Col-0_rosette-leaves | GEO | Arabidopsis thaliana | GSE22276 | Download (0) | |
| H3 | Col-0 | Col-0_old-leaves | 4w | GEO | Arabidopsis thaliana | GSE36629 | Download (5,156) |
| H3 | Col-0 | Col-0_meristem-young-leaves | 4w | GEO | Arabidopsis thaliana | GSE36629 | Download (9,961) |
| H3 | Col-x-Ler | Col-x-Ler_endosperm | 20d-2 | GEO | Arabidopsis thaliana | GSE66585 | Download (14,826) |
| H3 | Ler-x-Col | Ler-x-Col_endosperm | 20d-1 | GEO | Arabidopsis thaliana | GSE66585 | Download (11,181) |
| H3 | Col-0 | Col-0_flower | 5w-mutant | GEO | Arabidopsis thaliana | GSE106942 | Download (33,739) |
| H3 | Ler-x-Col | Ler-x-Col_endosperm | 20d-2 | GEO | Arabidopsis thaliana | GSE66585 | Download (2,893) |
| H3 | Col-0 | Col-0_leaves-apical-meristem | 5w-16c | GEO | Arabidopsis thaliana | GSE85279 | Download (33,419) |
| H3 | Col-0 | Col-0_leaves-apical-meristem | 5w-25c | GEO | Arabidopsis thaliana | GSE85279 | Download (32,878) |
| H3-1 | Col-0 | Col-0_meristem-young-leaves | 4w | GEO | Arabidopsis thaliana | GSE36629 | Download (6,011) |
| H3-1 | Col-0 | Col-0_old-leaves | 4w | GEO | Arabidopsis thaliana | GSE36629 | Download (3,432) |
| H3-1 | Col-0 | Col-0_seedling | 10d | GEO | Arabidopsis thaliana | GSE34840 | Download (27,065) |
| H3-1 | Col-0 | Col-0_leaves | 3w-A31T | GEO | Arabidopsis thaliana | GSE93223 | Download (9,522) |
| H3-3 | Col-0 | Col-0_seedling | 10d | GEO | Arabidopsis thaliana | GSE34840 | Download (27,811) |
| H3-3 | Col-0 | Col-0_whole-plant | 2w-wt | GEO | Arabidopsis thaliana | GSE87917 | Download (13,983) |
| H3-3 | Col-0 | Col-0_leaves | 3w-swap | GEO | Arabidopsis thaliana | GSE93223 | Download (32,660) |
| H3-3 | Col-0 | Col-0_leaves | 3w-T31A | GEO | Arabidopsis thaliana | GSE93223 | Download (30,864) |
| H3-3 | Col-0 | Col-0_meristem-young-leaves | 4w | GEO | Arabidopsis thaliana | GSE36629 | Download (15,168) |
| H3-3 | Col-0 | Col-0_old-leaves | 4w | GEO | Arabidopsis thaliana | GSE36629 | Download (12,319) |
| H3K14ac | Col-0 | Col-0_seedling | 3d-ein2-air | GEO | Arabidopsis thaliana | GSE77394 | Download (6,729) |
| H3K14ac | Col-0 | Col-0_seedling | 3d-ein2-ehylene | GEO | Arabidopsis thaliana | GSE77394 | Download (3,171) |
| H3K14ac | Col-0 | Col-0_seedling | 3d-wt-air | GEO | Arabidopsis thaliana | GSE77394 | Download (194) |
| H3K14ac | Col-0 | Col-0_seedling | 3d-wt-ehylene | GEO | Arabidopsis thaliana | GSE77394 | Download (12,779) |
| H3K14ac | Col-0 | Col-0_seedling | 12d-acc1 | GEO | Arabidopsis thaliana | GSE79524 | Download (16,324) |
| H3K14ac | Col-0 | Col-0_seedling | 12d-wt | GEO | Arabidopsis thaliana | GSE79524 | Download (15,816) |
| H3K14ac | Col-0 | Col-0_seedling | 10d-hdac | GEO | Arabidopsis thaliana | GSE89768 | Download (16,919) |
| H3K14ac | Col-0 | Col-0_seedling | 10d-pwr2 | GEO | Arabidopsis thaliana | GSE89768 | Download (16,568) |
| H3K14ac | Col-0 | Col-0_seedling | 10d-wt | GEO | Arabidopsis thaliana | GSE89768 | Download (17,336) |
| H3K18ac | Col-0 | Col-0_seedling | 12d-acc1 | GEO | Arabidopsis thaliana | GSE79524 | Download (12,299) |
| H3K18ac | Col-0 | Col-0_seedling | 12d-wt | GEO | Arabidopsis thaliana | GSE79524 | Download (10,484) |
| H3K23ac | Col-0 | Col-0_seedling | 3d-ein2-air | GEO | Arabidopsis thaliana | GSE77394 | Download (9,451) |
| H3K23ac | Col-0 | Col-0_seedling | 3d-ein2-ehylene | GEO | Arabidopsis thaliana | GSE77394 | Download (5,956) |
| H3K23ac | Col-0 | Col-0_seedling | 3d-wt-air | GEO | Arabidopsis thaliana | GSE77394 | Download (1,299) |
| H3K23ac | Col-0 | Col-0_seedling | 3d-wt-ehylene | GEO | Arabidopsis thaliana | GSE77394 | Download (9,899) |
| H3K27ac | Col-0 | Col-0_seedling | 12d-2xmutant-induced | GEO | Arabidopsis thaliana | GSE79524 | Download (16,971) |
| H3K27ac | Col-0 | Col-0_seedling | 12d-2xmutant-mock | GEO | Arabidopsis thaliana | GSE79524 | Download (16,072) |
| H3K27ac | Col-0 | Col-0_seedling | 12d-acc1 | GEO | Arabidopsis thaliana | GSE79524 | Download (15,534) |
| H3K27ac | Col-0 | Col-0_seedling | 12d-wt | GEO | Arabidopsis thaliana | GSE79524 | Download (14,145) |
| H3K27ac | Col-0 | Col-0_mature-early-senescent-leaves | hda9 | GEO | Arabidopsis thaliana | GSE80056 | Download (13,832) |
| H3K27ac | Col-0 | Col-0_mature-early-senescent-leaves | pwr | GEO | Arabidopsis thaliana | GSE80056 | Download (13,554) |
| H3K27ac | Col-0 | Col-0_mature-early-senescent-leaves | wt | GEO | Arabidopsis thaliana | GSE80056 | Download (10,021) |
| H3K27me1 | Col-x-Ler | Col-x-Ler_endosperm | 20d | GEO | Arabidopsis thaliana | GSE66585 | Download (13,747) |
| H3K27me1 | Ler-x-Col | Ler-x-Col_endosperm | 20d-2 | GEO | Arabidopsis thaliana | GSE66585 | Download (12,334) |
| H3K27me1 | Col-0 | Col-0_rosette-leaves | GEO | Arabidopsis thaliana | GSE54894 | Download (28,242) | |
| H3K27me1 | Col-0 | Col-0_leaves | 20d | GEO | Arabidopsis thaliana | GSE66585 | Download (5,694) |
| H3K27me3 | Col-0 | Col-0_seedling | 10d-clf | GEO | Arabidopsis thaliana | GSE108960 | Download (6,001) |
| H3K27me3 | Col-0 | Col-0_seedling | 10d-swn | GEO | Arabidopsis thaliana | GSE108960 | Download (6,433) |
| H3K27me3 | Col-0 | Col-0_seedling | 10d-wt | GEO | Arabidopsis thaliana | GSE108960 | Download (6,581) |
| H3K27me3 | Col-0 | Col-0_seedling | 10d-h3-1kd-1 | GEO | Arabidopsis thaliana | GSE101220 | Download (8,967) |
| H3K27me3 | Col-0 | Col-0_seedling | 10d-h3-1kd-2 | GEO | Arabidopsis thaliana | GSE101220 | Download (9,275) |
| H3K27me3 | Col-0 | Col-0_seedling | 10d-wt | GEO | Arabidopsis thaliana | GSE101220 | Download (9,505) |
| H3K27me3 | Col-0 | Col-0_seedling | 10d-ref6 | GEO | Arabidopsis thaliana | GSE25446 | Download (10,000) |
| H3K27me3 | Col-0 | Col-0_seedling | 10d-wt | GEO | Arabidopsis thaliana | GSE25446 | Download (9,776) |
| H3K27me3 | Col-0 | Col-0_seedling | 14d | GEO | Arabidopsis thaliana | GSE42695 | Download (14,319) |
| H3K27me3 | Col-0 | Col-0_seedling | 14d-brm | GEO | Arabidopsis thaliana | GSE47202 | Download (7,569) |
| H3K27me3 | Col-0 | Col-0_seedling | 14d-wt | GEO | Arabidopsis thaliana | GSE47202 | Download (7,886) |
| H3K27me3 | Col-0 | Col-0_seedling | 14d-brm | GEO | Arabidopsis thaliana | GSE53620 | Download (7,175) |
| H3K27me3 | Col-0 | Col-0_seedling | 14d-brm-clf | GEO | Arabidopsis thaliana | GSE53620 | Download (7,329) |
| H3K27me3 | Col-0 | Col-0_seedling | 14d-clf | GEO | Arabidopsis thaliana | GSE53620 | Download (7,246) |
| H3K27me3 | Col-0 | Col-0_seedling | 14d-wt | GEO | Arabidopsis thaliana | GSE53620 | Download (7,204) |
| H3K27me3 | Col-0 | Col-0_seedling | 14d-clf | GEO | Arabidopsis thaliana | GSE62615 | Download (8,209) |
| H3K27me3 | Col-0 | Col-0_seedling | 14d-wt | GEO | Arabidopsis thaliana | GSE62615 | Download (10,109) |
| H3K27me3 | Col-0 | Col-0_seedling | 12d-acc1 | GEO | Arabidopsis thaliana | GSE79524 | Download (7,624) |
| H3K27me3 | Col-0 | Col-0_seedling | 12d-wt | GEO | Arabidopsis thaliana | GSE79524 | Download (7,076) |
| H3K27me3 | Col-0 | Col-0_seedling | 7d-atbmi | GEO | Arabidopsis thaliana | GSE89357 | Download (7,012) |
| H3K27me3 | Col-0 | Col-0_seedling | 7d-clf-swn | GEO | Arabidopsis thaliana | GSE89357 | Download (7,356) |
| H3K27me3 | Col-0 | Col-0_seedling | 7d-wt | GEO | Arabidopsis thaliana | GSE89357 | Download (7,356) |
| H3K27me3 | C24 | C24_seedling | 14d-clf | GEO | Arabidopsis thaliana | GSE62615 | Download (7,985) |
| H3K27me3 | C24 | C24_seedling | 14d-wt | GEO | Arabidopsis thaliana | GSE62615 | Download (10,255) |
| H3K27me3 | C24 | C24_seedling | 10d-pold2 | GEO | Arabidopsis thaliana | GSE79259 | Download (16,216) |
| H3K27me3 | C24 | C24_seedling | 10d-fen1 | GEO | Arabidopsis thaliana | GSE79738 | Download (16,031) |
| H3K27me3 | C24 | C24_seedling | 10d-wt | GEO | Arabidopsis thaliana | GSE79738 | Download (14,404) |
| H3K27me3 | Col-0 | Col-0_whole-plant | 30h | GEO | Arabidopsis thaliana | GSE84483 | Download (9,719) |
| H3K27me3 | Col-0 | Col-0_leaves | 20d | GEO | Arabidopsis thaliana | GSE66585 | Download (6,944) |
| H3K27me3 | Col-0 | Col-0_leaves | GEO | Arabidopsis thaliana | GSE75071 | Download (8,902) | |
| H3K27me3 | Col-0 | Col-0_roots | 14d-OTU5 | GEO | Arabidopsis thaliana | GSE81407 | Download (908) |
| H3K27me3 | Col-0 | Col-0_roots | 14d-wt | GEO | Arabidopsis thaliana | GSE81407 | Download (1,846) |
| H3K27me3 | Col-0 | Col-0_roots | 5d-mutant | GEO | Arabidopsis thaliana | GSE86429 | Download (8,645) |
| H3K27me3 | Col-0 | Col-0_roots | 5d-wt | GEO | Arabidopsis thaliana | GSE86429 | Download (10,358) |
| H3K27me3 | Col-0 | Col-0_aerial-part | 3w-clf-28 | GEO | Arabidopsis thaliana | GSE103361 | Download (10,694) |
| H3K27me3 | Col-0 | Col-0_aerial-part | 3w-pie1-5 | GEO | Arabidopsis thaliana | GSE103361 | Download (10,095) |
| H3K27me3 | Col-0 | Col-0_aerial-part | 3w-pkl-1 | GEO | Arabidopsis thaliana | GSE103361 | Download (11,980) |
| H3K27me3 | Col-0 | Col-0_aerial-part | 3w-wt | GEO | Arabidopsis thaliana | GSE103361 | Download (9,232) |
| H3K27me3 | Col-0 | Col-0_aerial-part | 6d-caa39 | GEO | Arabidopsis thaliana | GSE103924 | Download (8,395) |
| H3K27me3 | Col-0 | Col-0_aerial-part | 6d-wt | GEO | Arabidopsis thaliana | GSE103924 | Download (8,361) |
| H3K27me3 | Ler | Ler_partial-inflorescence | 10d-mutant | GEO | Arabidopsis thaliana | GSE71564 | Download (11,310) |
| H3K27me3 | Col-x-C24 | Col-x-C24_seedling | 14d-clf | GEO | Arabidopsis thaliana | GSE62615 | Download (8,703) |
| H3K27me3 | Col-x-C24 | Col-x-C24_seedling | 14d-wt | GEO | Arabidopsis thaliana | GSE62615 | Download (10,649) |
| H3K27me3 | Col-0 | Col-0_flower | 5w-wt | GEO | Arabidopsis thaliana | GSE106942 | Download (10,065) |
| H3K27me3 | Col-x-Ler | Col-x-Ler_endosperm | 20d | GEO | Arabidopsis thaliana | GSE66585 | Download (11,433) |
| H3K27me3 | Col-0 | Col-0_flower | 5w-mutant | GEO | Arabidopsis thaliana | GSE106942 | Download (12,322) |
| H3K27me3 | Col-0 | Col-0_aerial-part | 2w-wt | GEO | Arabidopsis thaliana | GSE67322 | Download (4,609) |
| H3K27me3 | Col-0 | Col-0_aerial-part | 2w-atbmi | GEO | Arabidopsis thaliana | GSE67322 | Download (6,629) |
| H3K27me3 | Ler | Ler_partial-inflorescence | 10d-mutant-Dex | GEO | Arabidopsis thaliana | GSE71564 | Download (8,508) |
| H3K27me3 | Ler | Ler_whole-inflorescence | 10d-wt | GEO | Arabidopsis thaliana | GSE71564 | Download (8,957) |
| H3K27me3 | Col-0 | Col-0_aerial-part | 2w-tfl | GEO | Arabidopsis thaliana | GSE67322 | Download (6,036) |
| H3K27me3 | Col-0 | Col-0_aerial-part | 2w-lhp | GEO | Arabidopsis thaliana | GSE67322 | Download (10,313) |
| H3K27me3 | Ler | Ler_rosette-leaves | 10d-wt | GEO | Arabidopsis thaliana | GSE71564 | Download (10,908) |
| H3K27me3 | Col-0 | Col-0_aerial-part | 2w-clf | GEO | Arabidopsis thaliana | GSE67322 | Download (1,225) |
| H3K27me3 | Col-0 | Col-0_aerial-part | 2w-atring | GEO | Arabidopsis thaliana | GSE67322 | Download (9,416) |
| H3K27me3 | Col-0 | Col-0_rosette-leaves | GEO | Arabidopsis thaliana | GSE50636 | Download (9,809) | |
| H3K36ac | Col-0 | Col-0_leaves | 35d-sdg8-Xchip | GEO | Arabidopsis thaliana | GSE74841 | Download (16,287) |
| H3K36ac | Col-0 | Col-0_leaves | 35d-wt-Xchip | GEO | Arabidopsis thaliana | GSE74841 | Download (16,386) |
| H3K36me3 | Col-0 | Col-0_seedling | 10d-H3-KD | GEO | Arabidopsis thaliana | GSE96834 | Download (15,924) |
| H3K36me3 | Col-0 | Col-0_seedling | 10d-wt | GEO | Arabidopsis thaliana | GSE96834 | Download (15,594) |
| H3K36me3 | Col-0 | Col-0_leaves | GEO | Arabidopsis thaliana | GSE75071 | Download (13,352) | |
| H3K36me3 | Col-0 | Col-0_leaves | 3w-K36M-heteroz | GEO | Arabidopsis thaliana | GSE92449 | Download (8,866) |
| H3K36me3 | Col-0 | Col-0_leaves | 3w-K36M-homoz | GEO | Arabidopsis thaliana | GSE92449 | Download (7,115) |
| H3K36me3 | Col-0 | Col-0_leaves | 3w-sdg8-homoz | GEO | Arabidopsis thaliana | GSE92449 | Download (10,531) |
| H3K36me3 | Col-0 | Col-0_leaves | 3w-transgenic-H3 | GEO | Arabidopsis thaliana | GSE92449 | Download (13,343) |
| H3K36me3 | Col-0 | Col-0_leaves | 3w-wt | GEO | Arabidopsis thaliana | GSE92449 | Download (13,335) |
| H3K36me3 | Ws | Ws_leaves | 35d-wt-Xchip | GEO | Arabidopsis thaliana | GSE74841 | Download (12,265) |
| H3K36me3 | Ws | Ws_leaves | 35d-gcn5-Xchip | GEO | Arabidopsis thaliana | GSE74841 | Download (11,750) |
| H3K36me3 | Col-0 | Col-0_leaves-apical-meristem | 5w-25c | GEO | Arabidopsis thaliana | GSE85279 | Download (15,620) |
| H3K36me3 | Col-0 | Col-0_leaves-apical-meristem | 5w-16c | GEO | Arabidopsis thaliana | GSE85279 | Download (15,774) |
| H3K4me1 | Col-0 | Col-0_rosette-leaves | 21d-mutant-drought | GEO | Arabidopsis thaliana | GSE68370 | Download (28,889) |
| H3K4me1 | Col-0 | Col-0_rosette-leaves | 21d-wt-watered | GEO | Arabidopsis thaliana | GSE68370 | Download (27,712) |
| H3K4me1 | Col-0 | Col-0_rosette-leaves | 21d-wt-drought | GEO | Arabidopsis thaliana | GSE68370 | Download (28,257) |
| H3K4me1 | Col-0 | Col-0_rosette-leaves | 21d-mutant-watered | GEO | Arabidopsis thaliana | GSE68370 | Download (26,911) |
| H3K4me3 | Col-0 | Col-0_rosette-leaves | 21d-wt-watered | GEO | Arabidopsis thaliana | GSE68370 | Download (28,635) |
| H3K4me3 | Ler | Ler_partial-inflorescence | 10d-mutant | GEO | Arabidopsis thaliana | GSE71564 | Download (16,881) |
| H3K4me3 | Ler | Ler_partial-inflorescence | 10d-mutant-Dex | GEO | Arabidopsis thaliana | GSE71564 | Download (16,108) |
| H3K4me3 | Ler | Ler_whole-inflorescence | 10d-wt | GEO | Arabidopsis thaliana | GSE71564 | Download (16,422) |
| H3K4me3 | Col-0 | Col-0_rosette-leaves | 57d | GEO | Arabidopsis thaliana | GSE67776 | Download (16,706) |
| H3K4me3 | Ler | Ler_rosette-leaves | 10d-wt | GEO | Arabidopsis thaliana | GSE71564 | Download (16,907) |
| H3K4me3 | Col-0 | Col-0_rosette-leaves | 42d | GEO | Arabidopsis thaliana | GSE67776 | Download (16,100) |
| H3K4me3 | Col-0 | Col-0_rosette-leaves | 35d | GEO | Arabidopsis thaliana | GSE67776 | Download (16,138) |
| H3K4me3 | Col-0 | Col-0_rosette-leaves | 29d | GEO | Arabidopsis thaliana | GSE67776 | Download (15,277) |
| H3K4me3 | Col-0 | Col-0_rosette-leaves | 21d-mutant-drought | GEO | Arabidopsis thaliana | GSE68370 | Download (26,611) |
| H3K4me3 | Col-0 | Col-0_rosette-leaves | GEO | Arabidopsis thaliana | GSE50636 | Download (16,484) | |
| H3K4me3 | Col-0 | Col-0_rosette-leaves | GEO | Arabidopsis thaliana | GSE22276 | Download (0) | |
| H3K4me3 | Col-0 | Col-0_rosette-leaves | 21d-mutant-watered | GEO | Arabidopsis thaliana | GSE68370 | Download (26,820) |
| H3K4me3 | Col-0 | Col-0_rosette-leaves | 21d-wt-drought | GEO | Arabidopsis thaliana | GSE68370 | Download (26,448) |
| H3K4me3 | Col-0 | Col-0_seedling | 10d-H3-KD | GEO | Arabidopsis thaliana | GSE96834 | Download (22,559) |
| H3K4me3 | Col-0 | Col-0_seedling | 10d-wt | GEO | Arabidopsis thaliana | GSE96834 | Download (23,219) |
| H3K4me3 | C24 | C24_seedling | 10d-pold2 | GEO | Arabidopsis thaliana | GSE79259 | Download (17,314) |
| H3K4me3 | C24 | C24_seedling | 10d-wt | GEO | Arabidopsis thaliana | GSE79259 | Download (16,815) |
| H3K4me3 | C24 | C24_seedling | 10d-fen1 | GEO | Arabidopsis thaliana | GSE79738 | Download (17,002) |
| H3K4me3 | C24 | C24_seedling | 10d-wt | GEO | Arabidopsis thaliana | GSE79738 | Download (16,881) |
| H3K4me3 | Col-0 | Col-0_leaves | GEO | Arabidopsis thaliana | GSE75071 | Download (17,726) | |
| H3K4me3 | Col-0 | Col-0_roots | 14d-OTU5 | GEO | Arabidopsis thaliana | GSE81407 | Download (2,593) |
| H3K4me3 | Col-0 | Col-0_roots | 14d-wt | GEO | Arabidopsis thaliana | GSE81407 | Download (6,193) |
| H3K4me3 | Col-0 | Col-0_roots | 5d-mutant | GEO | Arabidopsis thaliana | GSE86429 | Download (16,389) |
| H3K4me3 | Col-0 | Col-0_roots | 5d-mutant-trimmed | GEO | Arabidopsis thaliana | GSE86429 | Download (1,542) |
| H3K4me3 | Col-0 | Col-0_roots | 5d-wt | GEO | Arabidopsis thaliana | GSE86429 | Download (11,370) |
| H3K56ac | Col-0 | Col-0_leaves | GEO | Arabidopsis thaliana | GSE75071 | Download (17,561) | |
| H3K9-14ac | Col-0 | Col-0_seedling | 7d-GSH-3h | GEO | Arabidopsis thaliana | GSE82075 | Download (16,426) |
| H3K9-14ac | Col-0 | Col-0_seedling | 7d-GSH-6h | GEO | Arabidopsis thaliana | GSE82075 | Download (17,154) |
| H3K9-14ac | Col-0 | Col-0_seedling | 7d-GSNO-3h | GEO | Arabidopsis thaliana | GSE82075 | Download (15,829) |
| H3K9-14ac | Col-0 | Col-0_seedling | 7d-GSNO-6h | GEO | Arabidopsis thaliana | GSE82075 | Download (17,619) |
| H3K9-14ac | Col-0 | Col-0_seedling | 7d-GSNO-cPTIO-3h | GEO | Arabidopsis thaliana | GSE82075 | Download (16,818) |
| H3K9-14ac | Col-0 | Col-0_seedling | 7d-GSNO-cPTIO-6h | GEO | Arabidopsis thaliana | GSE82075 | Download (18,089) |
| H3K9-14ac | Col-0 | Col-0_seedling | 7d-TSA-3h | GEO | Arabidopsis thaliana | GSE82075 | Download (17,902) |
| H3K9-14ac | Col-0 | Col-0_seedling | 7d-TSA-6h | GEO | Arabidopsis thaliana | GSE82075 | Download (16,196) |
| H3K9-14ac | Col-0 | Col-0_seedling | 7d-water-3h | GEO | Arabidopsis thaliana | GSE82075 | Download (16,902) |
| H3K9-14ac | Col-0 | Col-0_seedling | 7d-water-6h | GEO | Arabidopsis thaliana | GSE82075 | Download (16,452) |
| H3K9ac | Col-0 | Col-0_seedling | 3d-wt-ehylene | GEO | Arabidopsis thaliana | GSE77394 | Download (15,691) |
| H3K9ac | Col-0 | Col-0_seedling | 12d-acc1 | GEO | Arabidopsis thaliana | GSE79524 | Download (16,223) |
| H3K9ac | Col-0 | Col-0_seedling | 12d-wt | GEO | Arabidopsis thaliana | GSE79524 | Download (15,826) |
| H3K9ac | Col-0 | Col-0_seedling | 10d-hdac | GEO | Arabidopsis thaliana | GSE89768 | Download (16,205) |
| H3K9ac | Col-0 | Col-0_seedling | 10d-pwr2 | GEO | Arabidopsis thaliana | GSE89768 | Download (15,592) |
| H3K9ac | Col-0 | Col-0_seedling | 10d-wt | GEO | Arabidopsis thaliana | GSE89768 | Download (17,957) |
| H3K9ac | Col-0 | Col-0_seedling | 3d-air | GEO | Arabidopsis thaliana | GSE93875 | Download (15,979) |
| H3K9ac | Col-0 | Col-0_seedling | 3d-C2H4 | GEO | Arabidopsis thaliana | GSE93875 | Download (9,325) |
| H3K9ac | Col-0 | Col-0_seedling | 14d-flg22 | GEO | Arabidopsis thaliana | GSE99936 | Download (17,890) |
| H3K9ac | Col-0 | Col-0_seedling | 14d-mock | GEO | Arabidopsis thaliana | GSE99936 | Download (18,456) |
| H3K9ac | Col-0 | Col-0_rosette-leaves | GEO | Arabidopsis thaliana | GSE22276 | Download (0) | |
| H3K9ac | Col-0 | Col-0_rosette-leaves | 42d | GEO | Arabidopsis thaliana | GSE67776 | Download (14,700) |
| H3K9ac | Col-0 | Col-0_rosette-leaves | 34d | GEO | Arabidopsis thaliana | GSE67776 | Download (15,507) |
| H3K9ac | Col-0 | Col-0_rosette-leaves | 30d | GEO | Arabidopsis thaliana | GSE67776 | Download (15,530) |
| H3K9me2 | Ler-x-Col | Ler-x-Col_endosperm | 20d | GEO | Arabidopsis thaliana | GSE66585 | Download (8,668) |
| H3K9me2 | Col-x-Ler | Col-x-Ler_endosperm | 20d | GEO | Arabidopsis thaliana | GSE66585 | Download (7,738) |
| H3K9me2 | Col-0 | Col-0_seedling | 12-14d-suvh1-3 | GEO | Arabidopsis thaliana | GSE108414 | Download (5,811) |
| H3K9me2 | Col-0 | Col-0_seedling | 12-14d-wt | GEO | Arabidopsis thaliana | GSE108414 | Download (5,854) |
| H3K9me2 | Col-0 | Col-0_seedling | 10d-atxr | GEO | Arabidopsis thaliana | GSE111811 | Download (6,966) |
| H3K9me2 | Col-0 | Col-0_seedling | 10d-se-2 | GEO | Arabidopsis thaliana | GSE111811 | Download (7,494) |
| H3K9me2 | Col-0 | Col-0_seedling | 10d-se-2-atxr | GEO | Arabidopsis thaliana | GSE111811 | Download (7,282) |
| H3K9me2 | Col-0 | Col-0_seedling | 10d-wt | GEO | Arabidopsis thaliana | GSE111811 | Download (6,552) |
| H3K9me2 | Col-0 | Col-0_seedling | 3w-crh6 | GEO | Arabidopsis thaliana | GSE37644 | Download (17,540) |
| H3K9me2 | Col-0 | Col-0_seedling | 3w-crt1 | GEO | Arabidopsis thaliana | GSE37644 | Download (23,805) |
| H3K9me2 | Col-0 | Col-0_seedling | 3w-wt | GEO | Arabidopsis thaliana | GSE37644 | Download (28,945) |
| H3K9me2 | C24 | C24_seedling | 10d-pold2 | GEO | Arabidopsis thaliana | GSE79259 | Download (6,870) |
| H3K9me2 | C24 | C24_seedling | 10d-wt | GEO | Arabidopsis thaliana | GSE79259 | Download (6,654) |
| H3K9me2 | Col-0 | Col-0_endosperm | 4d-wt-x-wt | GEO | Arabidopsis thaliana | GSE100011 | Download (7,022) |
| H3K9me2 | Col-0 | Col-0_aerial-part | 6d-caa39 | GEO | Arabidopsis thaliana | GSE103924 | Download (5,034) |
| H3K9me2 | Col-0 | Col-0_aerial-part | 6d-wt | GEO | Arabidopsis thaliana | GSE103924 | Download (5,220) |
| H3T3ph | Col-0 | Col-0_rosette-leaves | 21d-mutant-drought | GEO | Arabidopsis thaliana | GSE68370 | Download (30,935) |
| H3T3ph | Col-0 | Col-0_rosette-leaves | 21d-mutant-watered | GEO | Arabidopsis thaliana | GSE68370 | Download (31,193) |
| H3T3ph | Col-0 | Col-0_rosette-leaves | 21d-wt-drought | GEO | Arabidopsis thaliana | GSE68370 | Download (32,193) |
| H3T3ph | Col-0 | Col-0_rosette-leaves | 21d-wt-watered | GEO | Arabidopsis thaliana | GSE68370 | Download (33,262) |
| H3ac | Col-0 | Col-0_leaves | 4w-hac1-5 | GEO | Arabidopsis thaliana | GSE101570 | Download (15,782) |
| H3ac | Col-0 | Col-0_leaves | 4w-hac1-5-INA | GEO | Arabidopsis thaliana | GSE101570 | Download (16,001) |
| H3ac | Col-0 | Col-0_leaves | 4w-npr1-1 | GEO | Arabidopsis thaliana | GSE101570 | Download (15,987) |
| H3ac | Col-0 | Col-0_leaves | 4w-npr1-1-INA | GEO | Arabidopsis thaliana | GSE101570 | Download (16,398) |
| H3ac | Col-0 | Col-0_leaves | 4w-wt | GEO | Arabidopsis thaliana | GSE101570 | Download (16,234) |
| H3ac | Col-0 | Col-0_leaves | 4w-wt-INA | GEO | Arabidopsis thaliana | GSE101570 | Download (16,294) |
| H3ac | Col-0 | Col-0_roots | 7d-hag1 | GEO | Arabidopsis thaliana | GSE100965 | Download (16,270) |
| H3ac | Col-0 | Col-0_roots | 7d-wt | GEO | Arabidopsis thaliana | GSE100965 | Download (16,566) |
| H4K12ac | Col-0 | Col-0_seedling | 12d-acc1 | GEO | Arabidopsis thaliana | GSE79524 | Download (17,414) |
| H4K12ac | Col-0 | Col-0_seedling | 12d-wt | GEO | Arabidopsis thaliana | GSE79524 | Download (16,455) |
| H4K16ac | Col-0 | Col-0_seedling | 12d-acc1 | GEO | Arabidopsis thaliana | GSE79524 | Download (17,314) |
| H4K16ac | Col-0 | Col-0_seedling | 12d-wt | GEO | Arabidopsis thaliana | GSE79524 | Download (16,782) |
| H4K5ac | Col-0 | Col-0_seedling | 12d-acc1 | GEO | Arabidopsis thaliana | GSE79524 | Download (17,777) |
| H4K5ac | Col-0 | Col-0_seedling | 12d-wt | GEO | Arabidopsis thaliana | GSE79524 | Download (17,235) |
| H4K5ac | undef | undef_seedling | 10d-wt | GEO | Arabidopsis thaliana | GSE116065 | Download (30,951) |
| H4K5ac | undef | undef_seedling | 10d-epcr | GEO | Arabidopsis thaliana | GSE116065 | Download (23,444) |
| H4K5ac | undef | undef_seedling | 10d-arid | GEO | Arabidopsis thaliana | GSE116065 | Download (26,898) |
| H4K8ac | Col-0 | Col-0_seedling | 12d-acc1 | GEO | Arabidopsis thaliana | GSE79524 | Download (18,148) |
| H4K8ac | Col-0 | Col-0_seedling | 12d-wt | GEO | Arabidopsis thaliana | GSE79524 | Download (17,324) |
| H4ac | Col-0 | Col-0_seedling | 6d-arp | GEO | Arabidopsis thaliana | GSE117391 | Download (14,240) |
| H4ac | Col-0 | Col-0_seedling | 6d-mbd | GEO | Arabidopsis thaliana | GSE117391 | Download (13,727) |
| H4ac | Col-0 | Col-0_seedling | 6d-wt | GEO | Arabidopsis thaliana | GSE117391 | Download (14,735) |
| HAND2 | Kelly | GEO | Homo sapiens | GSE94822 | Download (34,946) | ||
| HAND2 | SK-N-BE2-C | GEO | Homo sapiens | GSE94822 | Download (78,820) | ||
| HAND2 | CLB-Ga | GEO | Homo sapiens | GSE90683 | Download (57,260) | ||
| HAT2 | Col-0 | Col-0_leaves | ecoli_col | GEO | Arabidopsis thaliana | GSE60141 | Download (506) |
| HAT22 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (18,250) |
| HAT22 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (15,811) |
| HAT3 | Col-0 | Col-0_whole-plant | 10d | GEO | Arabidopsis thaliana | GSE65876 | Download (140) |
| HAT5 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (119) |
| HB-5 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (285) |
| HBP1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR616WEG | Download (31,612) | ||
| HCFC1 | GM12878 | ENCODE | Homo sapiens | ENCSR514VAY | Download (33,149) | ||
| HCFC1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR529JYA | Download (17,532) | ||
| HCFC1 | K-562 | ENCODE | Homo sapiens | ENCSR000EFN | Download (26,682) | ||
| HCFC1 | MCF-7 | ENCODE | Homo sapiens | ENCSR697CUP | Download (12,907) | ||
| HCFC1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECH | Download (14,277) | ||
| HCFC1 | HeLa | GEO | Homo sapiens | GSE31417 | Download (1,885) | ||
| HCFC1R1 | cartilage | GEO | Homo sapiens | GSE100311 | Download (16,614) | ||
| HDAC1 | K-562 | ENCODE | Homo sapiens | ENCSR711VWL | Download (56,730) | ||
| HDAC1 | VCaP | DHAT_2H | GEO | Homo sapiens | GSE28950 | Download (4,022) | |
| HDAC1 | VCaP | ETOH | GEO | Homo sapiens | GSE28950 | Download (3,086) | |
| HDAC1 | K-562 | ENCODE | Homo sapiens | ENCSR000AQF | Download (21,267) | ||
| HDAC1 | K-562 | ENCODE | Homo sapiens | ENCSR387UWP | Download (61,074) | ||
| HDAC1 | K-562 | ENCODE | Homo sapiens | ENCSR568PGX | Download (806) | ||
| HDAC2 | GM12878 | ENCODE | Homo sapiens | ENCSR330OEO | Download (7,945) | ||
| HDAC2 | A-549 | ENCODE | Homo sapiens | ENCSR659LJJ | Download (10,247) | ||
| HDAC2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BMC | Download (68,960) | ||
| HDAC2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR337NWW | Download (5,313) | ||
| HDAC2 | K-562 | ENCODE | Homo sapiens | ENCSR000AQG | Download (14,956) | ||
| HDAC2 | K-562 | ENCODE | Homo sapiens | ENCSR000BMG | Download (21,812) | ||
| HDAC2 | K-562 | ENCODE | Homo sapiens | ENCSR075HTM | Download (59,131) | ||
| HDAC2 | K-562 | ENCODE | Homo sapiens | ENCSR893WSB | Download (74,885) | ||
| HDAC2 | MCF-7 | ENCODE | Homo sapiens | ENCSR000BTP | Download (49,044) | ||
| HDAC2 | WA01 | ENCODE | Homo sapiens | ENCSR000AVB | Download (56,185) | ||
| HDAC2 | WA01 | ENCODE | Homo sapiens | ENCSR000BNR | Download (13,735) | ||
| HDAC2 | VCaP | DHAT_2H | GEO | Homo sapiens | GSE28950 | Download (10,120) | |
| HDAC2 | VCaP | ETOH | GEO | Homo sapiens | GSE28950 | Download (12,956) | |
| HDAC3 | VCaP | DHAT_2H | GEO | Homo sapiens | GSE28950 | Download (5,497) | |
| HDAC3 | VCaP | ETOH | GEO | Homo sapiens | GSE28950 | Download (4,029) | |
| HDAC3 | K-562 | ENCODE | Homo sapiens | ENCSR024LKA | Download (5,002) | ||
| HDAC6 | GM12878 | ENCODE | Homo sapiens | ENCSR933EYC | Download (11,219) | ||
| HDAC6 | K-562 | ENCODE | Homo sapiens | ENCSR000ATJ | Download (923) | ||
| HDAC6 | WA01 | ENCODE | Homo sapiens | ENCSR000ATQ | Download (1,392) | ||
| HDAC8 | K-562 | ENCODE | Homo sapiens | ENCSR835TCD | Download (23,567) | ||
| HDAC8 | K-562 | ENCODE | Homo sapiens | ENCSR000DJZ | Download (10,170) | ||
| HDG1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (748) |
| HDGF | GM12878 | ENCODE | Homo sapiens | ENCSR145XQO | Download (87,883) | ||
| HDGF | HEK293T | ENCODE | Homo sapiens | ENCSR522LDJ | Download (1,941) | ||
| HDGF | K-562 | ENCODE | Homo sapiens | ENCSR197ALX | Download (65,383) | ||
| HDGF | K-562 | ENCODE | Homo sapiens | ENCSR563YDA | Download (50,362) | ||
| HDGF | MCF-7 | ENCODE | Homo sapiens | ENCSR200CUA | Download (4,320) | ||
| HEC1 | Col-0 | Col-0_seedling | 12d | GEO | Arabidopsis thaliana | GSE94307 | Download (9,084) |
| HES1 | Hep-G2 | GEO | Homo sapiens | GSE97661 | Download (1,670) | ||
| HES1 | MCF-7 | ENCODE | Homo sapiens | ENCSR109ODF | Download (4,476) | ||
| HES1 | SW1353 | GEO | Homo sapiens | GSE60006 | Download (906) | ||
| HES1 | K-562 | ENCODE | Homo sapiens | ENCSR091JXL | Download (8,270) | ||
| HEXIM1 | HCT-116 | GEO | Homo sapiens | GSE72622 | Download (69,089) | ||
| HEXIM1 | A-375 | A771726 | GEO | Homo sapiens | GSE68052 | Download (1,701) | |
| HEXIM1 | A-375 | DMSO | GEO | Homo sapiens | GSE68052 | Download (5,884) | |
| HEXIM1-CDK9 | A-375 | GEO | Homo sapiens | GSE68047 | Download (5,868) | ||
| HHEX | Hep-G2 | ENCODE | Homo sapiens | ENCSR656JZL | Download (5,227) | ||
| HHO2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,829) |
| HHO2 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (1,143) |
| HHO3 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,727) |
| HHO3 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (2,087) |
| HHO5 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (5,318) |
| HHO5 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (3,862) |
| HHO6 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,650) |
| HHO6 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (12,939) |
| HIC1 | HEK293 | ENCODE | Homo sapiens | ENCSR803GYT | Download (29,130) | ||
| HIF1A | ccRCC | GEO | Homo sapiens | GSE86092 | Download (635) | ||
| HIF1A | RCC10 | GEO | Homo sapiens | GSE101063 | Download (5,076) | ||
| HIF1A | H1299 | GEO | Homo sapiens | GSE76770 | Download (2,664) | ||
| HIF1A | macrophage | HYPO | GEO | Homo sapiens | GSE43109 | Download (9,039) | |
| HIF1A | macrophage | HYPO_IL | GEO | Homo sapiens | GSE43109 | Download (2,091) | |
| HIF1A | macrophage | NORMO_IL | GEO | Homo sapiens | GSE43109 | Download (570) | |
| HIF1A | U2OS | DMOG | GEO | Homo sapiens | GSE85096 | Download (19,073) | |
| HIF1A | U2OS | DMSO | GEO | Homo sapiens | GSE85096 | Download (7,645) | |
| HIF1A | U2OS | trough_DMOG | GEO | Homo sapiens | GSE85096 | Download (27,511) | |
| HIF1A | U2OS | trough_DMSO | GEO | Homo sapiens | GSE85096 | Download (3,183) | |
| HIF1A | 786-O | GEO | Homo sapiens | GSE34871 | Download (2,552) | ||
| HIF1A | HUVEC-C | HYPOX | GEO | Homo sapiens | GSE39089 | Download (3,423) | |
| HIF1A | HUVEC-C | NOMO | GEO | Homo sapiens | GSE39089 | Download (339) | |
| HIF1A | T-47D | HAEGIN2 | GEO | Homo sapiens | GSE59935 | Download (278,415) | |
| HIF2A | ccRCC | GEO | Homo sapiens | GSE86092 | Download (3,315) | ||
| HINFP | K-562 | ENCODE | Homo sapiens | ENCSR619GFP | Download (3,451) | ||
| HKR1 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (158) | ||
| HLF | Hep-G2 | ENCODE | Homo sapiens | ENCSR528PSI | Download (37,846) | ||
| HMBOX1 | K-562 | ENCODE | Homo sapiens | ENCSR757IIU | Download (42,722) | ||
| HMBOX1 | HeLa | GEO | Homo sapiens | GSE46237 | Download (2,821) | ||
| HMBOX1 | WI-38VA13 | GEO | Homo sapiens | GSE46237 | Download (1,487) | ||
| HMG20A | Hep-G2 | ENCODE | Homo sapiens | ENCSR072GJV | Download (88,251) | ||
| HMGB1 | HUVEC-C | GEO | Homo sapiens | GSE98245 | Download (7,134) | ||
| HMGB1 | IMR-90 | GEO | Homo sapiens | GSE98245 | Download (1,384) | ||
| HMGB2 | HUVEC-C | GEO | Homo sapiens | GSE98245 | Download (17,657) | ||
| HMGB2 | IMR-90 | proliferating | GEO | Homo sapiens | GSE98245 | Download (6,819) | |
| HMGB2 | IMR-90 | senescent | GEO | Homo sapiens | GSE98245 | Download (782) | |
| HMGN3 | K-562 | ENCODE | Homo sapiens | ENCSR000DOB | Download (23,378) | ||
| HMGXB4 | Hep-G2 | ENCODE | Homo sapiens | ENCSR174GOO | Download (74,814) | ||
| HNF1A | Hep-G2 | ENCODE | Homo sapiens | ENCSR800QIT | Download (15,205) | ||
| HNF1A | HEE | 1 | GEO | Homo sapiens | GSE76376 | Download (15,543) | |
| HNF1A | HEE | 5 | GEO | Homo sapiens | GSE76376 | Download (9,513) | |
| HNF1A | NY15 | GEO | Homo sapiens | GSE108150 | Download (8,115) | ||
| HNF1A | NY8 | GEO | Homo sapiens | GSE108150 | Download (93,580) | ||
| HNF1B | H9 | ENA | Homo sapiens | ERP004206 | Download (2,760) | ||
| HNF1B | PDAC | GEO | Homo sapiens | GSE64557 | Download (29,225) | ||
| HNF4A | LoVo | PHASEM | GEO | Homo sapiens | GSE51290 | Download (1,507) | |
| HNF4A | LoVo | PHASES | GEO | Homo sapiens | GSE51290 | Download (11,657) | |
| HNF4A | liver | ENCODE | Homo sapiens | ENCSR445QRF | Download (107,053) | ||
| HNF4A | liver | ENCODE | Homo sapiens | ENCSR601OGE | Download (94,118) | ||
| HNF4A | liver | ENA | Homo sapiens | ERP002306 | Download (64,642) | ||
| HNF4A | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BLF | Download (62,632) | ||
| HNF4A | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEU | Download (21,163) | ||
| HNF4A | Hep-G2 | ENA | Homo sapiens | ERP000209 | Download (39,187) | ||
| HNF4A | CACO2 | DIFF | GEO | Homo sapiens | GSE23436 | Download (64,374) | |
| HNF4A | CACO2 | PROLIF | GEO | Homo sapiens | GSE23436 | Download (19,748) | |
| HNF4A | GP5D | GEO | Homo sapiens | GSE51234 | Download (14,885) | ||
| HNF4A | GP5D | SIRAD21 | GEO | Homo sapiens | GSE51234 | Download (4,148) | |
| HNF4A | HCT-116 | GEO | Homo sapiens | GSE62890 | Download (6,750) | ||
| HNF4A | HCT-116 | TCF4 | GEO | Homo sapiens | GSE62890 | Download (3,900) | |
| HNF4A | HCT-116 | TCF4_DOX | GEO | Homo sapiens | GSE62890 | Download (2,909) | |
| HNF4G | liver | ENCODE | Homo sapiens | ENCSR297GII | Download (21,951) | ||
| HNF4G | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BNJ | Download (38,973) | ||
| HNF4G | 22Rv1 | GEO | Homo sapiens | GSE85558 | Download (18,058) | ||
| HNF4G | 22Rv1 | Dox | GEO | Homo sapiens | GSE85558 | Download (16,355) | |
| HNRNPH1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR066FXN | Download (56,569) | ||
| HNRNPK | Hep-G2 | ENCODE | Homo sapiens | ENCSR519QAA | Download (51,273) | ||
| HNRNPK | K-562 | ENCODE | Homo sapiens | ENCSR014RCS | Download (20,192) | ||
| HNRNPL | Hep-G2 | ENCODE | Homo sapiens | ENCSR315JJE | Download (38,776) | ||
| HNRNPL | K-562 | ENCODE | Homo sapiens | ENCSR594BNR | Download (12,335) | ||
| HNRNPLL | Hep-G2 | ENCODE | Homo sapiens | ENCSR804HMZ | Download (52,099) | ||
| HNRNPLL | K-562 | ENCODE | Homo sapiens | ENCSR112RNT | Download (43,438) | ||
| HNRNPUL1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR183AXJ | Download (6,954) | ||
| HNRNPUL1 | K-562 | ENCODE | Homo sapiens | ENCSR296MXW | Download (4,558) | ||
| HOMEZ | Hep-G2 | ENCODE | Homo sapiens | ENCSR117CHD | Download (97,681) | ||
| HOXA9 | SEM | GEO | Homo sapiens | GSE38339 | Download (5,767) | ||
| HOXA9 | HEK293-FT | GEO | Homo sapiens | GSE62586 | Download (22,509) | ||
| HOXB13 | LNCaP | GEO | Homo sapiens | GSE56288 | Download (54,386) | ||
| HOXB13 | prostate | tumor_tissue1 | GEO | Homo sapiens | GSE114385 | Download (1,052) | |
| HOXB13 | prostate | tumor_tissue2 | GEO | Homo sapiens | GSE114385 | Download (760) | |
| HOXB13 | prostate | tumor_tissue3 | GEO | Homo sapiens | GSE114385 | Download (980) | |
| HOXB13 | prostate | tumor_tissue4 | GEO | Homo sapiens | GSE114385 | Download (1,303) | |
| HOXB13 | G-401 | GEO | Homo sapiens | GSE65381 | Download (14,253) | ||
| HOXB13 | 22Rv1 | GEO | Homo sapiens | GSE96652 | Download (11,594) | ||
| HOXB13 | LNCaP | GEO | Homo sapiens | GSE94682 | Download (94,492) | ||
| HOXB13 | LNCaP | r1881 | GEO | Homo sapiens | GSE94682 | Download (85,833) | |
| HOXB13 | LNCaP | GEO | Homo sapiens | GSE96652 | Download (36,283) | ||
| HOXB13 | VCaP | GEO | Homo sapiens | GSE96652 | Download (152,358) | ||
| HOXB13 | VCaP | siERG | GEO | Homo sapiens | GSE96652 | Download (160,095) | |
| HOXB7 | BT-474 | GEO | Homo sapiens | GSE47164 | Download (186) | ||
| HOXC11 | MCF-7 | LY2 | GEO | Homo sapiens | GSE54027 | Download (8) | |
| HOXC11 | MCF-7 | LY2_TAMOXIFEN | GEO | Homo sapiens | GSE54027 | Download (35) | |
| HOXC5 | PC-3 | GEO | Homo sapiens | GSE97570 | Download (84,072) | ||
| HOXC5 | PC-3 | Hoxc5overexp | GEO | Homo sapiens | GSE97570 | Download (2,982) | |
| HSF1 | NCI-H441 | GEO | Homo sapiens | GSE38901 | Download (1,182) | ||
| HSF1 | HCT-116 | GEO | Homo sapiens | GSE57398 | Download (23) | ||
| HSF1 | HCT-116 | KOFBXW7 | GEO | Homo sapiens | GSE57398 | Download (3,874) | |
| HSF1 | HCT-116 | KOFBXW7_HEAT | GEO | Homo sapiens | GSE57398 | Download (382) | |
| HSF1 | HME1 | GEO | Homo sapiens | GSE38901 | Download (909) | ||
| HSF1 | HME1 | HEAT | GEO | Homo sapiens | GSE38901 | Download (4,346) | |
| HSF1 | HMLER | GEO | Homo sapiens | GSE38901 | Download (233) | ||
| HSF1 | HT29 | GEO | Homo sapiens | GSE38901 | Download (1,783) | ||
| HSF1 | MO91 | GEO | Homo sapiens | GSE45852 | Download (28,737) | ||
| HSF1 | MO91 | 27A_100UM | GEO | Homo sapiens | GSE45852 | Download (1,623) | |
| HSF1 | MO91 | 27A_20UM | GEO | Homo sapiens | GSE45852 | Download (12,283) | |
| HSF1 | MO91 | CHX_10UM | GEO | Homo sapiens | GSE45852 | Download (19,650) | |
| HSF1 | SKBR3 | GEO | Homo sapiens | GSE38901 | Download (4,733) | ||
| HSF1 | SW620 | GEO | Homo sapiens | GSE38901 | Download (1,556) | ||
| HSF1 | U2OS | HEAT | GEO | Homo sapiens | GSE60984 | Download (8,928) | |
| HSF1 | U2OS | HEAT_20 | GEO | Homo sapiens | GSE60984 | Download (9,658) | |
| HSF1 | ZR751 | GEO | Homo sapiens | GSE38901 | Download (2,063) | ||
| HSF1 | BT-20 | GEO | Homo sapiens | GSE38901 | Download (3,669) | ||
| HSF1 | 451Lu | GEO | Homo sapiens | GSE63589 | Download (137,093) | ||
| HSF1 | MCF-10A | GEO | Homo sapiens | GSE38901 | Download (466) | ||
| HSF1 | MCF-10A | HEAT | GEO | Homo sapiens | GSE38901 | Download (4,218) | |
| HSF1 | MCF-7 | GEO | Homo sapiens | GSE38901 | Download (1,630) | ||
| HSF1 | MCF-7 | GEO | Homo sapiens | GSE45852 | Download (1,016) | ||
| HSF1 | MCF-7 | CHX_10UM | GEO | Homo sapiens | GSE45852 | Download (2,044) | |
| HSF1 | NCI-H1703 | GEO | Homo sapiens | GSE38901 | Download (446) | ||
| HSF1 | NCI-H838 | GEO | Homo sapiens | GSE38901 | Download (27,092) | ||
| HSF1 | P12 | GEO | Homo sapiens | GSE90716 | Download (267) | ||
| HSF1 | GM12878 | ENCODE | Homo sapiens | ENCSR009MBP | Download (7,607) | ||
| HSF1 | WA09 | heat-shock | GEO | Homo sapiens | GSE105028 | Download (1,620) | |
| HSF1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EET | Download (2,000) | ||
| HSF1 | MCF-7 | ENCODE | Homo sapiens | ENCSR062HDL | Download (648) | ||
| HSF1 | BPE | GEO | Homo sapiens | GSE38901 | Download (1,309) | ||
| HSF1 | BPE | HEAT | GEO | Homo sapiens | GSE38901 | Download (4,767) | |
| HSF1 | BPLER | GEO | Homo sapiens | GSE38901 | Download (2,689) | ||
| HSF1 | breast | tumor | GEO | Homo sapiens | GSE38901 | Download (3,738) | |
| HSF1 | colon | tumor | GEO | Homo sapiens | GSE38901 | Download (1,351) | |
| HSF1 | HCT15 | GEO | Homo sapiens | GSE38901 | Download (1,099) | ||
| HSFA1A | Col-0 | Col-0_seedling | 12d-15min-17c | GEO | Arabidopsis thaliana | GSE79354 | Download (1,516) |
| HSFA1A | Col-0 | Col-0_seedling | 12d-15min-27c | GEO | Arabidopsis thaliana | GSE79354 | Download (1,371) |
| HSFA1A | Col-0 | Col-0_seedling | 12d-15min-37c | GEO | Arabidopsis thaliana | GSE79354 | Download (1,434) |
| HSFA1B | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (4,624) |
| HSFA1B | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (13,608) |
| HSFA1B | Col-0 | Col-0_leaves | 5w-mutant-HS | GEO | Arabidopsis thaliana | GSE85651 | Download (345) |
| HSFA1B | Col-0 | Col-0_leaves | 5w-mutant-NS | GEO | Arabidopsis thaliana | GSE85651 | Download (12,695) |
| HSFA1B | Col-0 | Col-0_leaves | 5w-wt-HS | GEO | Arabidopsis thaliana | GSE85651 | Download (424) |
| HSFA1B | Col-0 | Col-0_leaves | 5w-wt-NS | GEO | Arabidopsis thaliana | GSE85651 | Download (119) |
| HSFA6A | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (7,368) |
| HSFA6A | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (232) |
| HSFA6A | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (332) |
| HSFA6B | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,372) |
| HSFB2A | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,094) |
| HSFB2B | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (254) |
| HSFB2B | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (1,899) |
| HSFB4 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (194) |
| HSFC1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,784) |
| HTA6 | Col-0 | Col-0_leaves | 2-3w | GEO | Arabidopsis thaliana | GSE95557 | Download (7,925) |
| HTR12 | Col-0 | Col-0_seedling | 7-10d-At | GEO | Arabidopsis thaliana | GSE88907 | Download (6,765) |
| HTR12 | Col-0 | Col-0_seedling | 7-10d-Lo | GEO | Arabidopsis thaliana | GSE88907 | Download (2,004) |
| HTR12 | Col-0 | Col-0_seedling | 7-10d-Zm | GEO | Arabidopsis thaliana | GSE88907 | Download (2,872) |
| HY5 | Ws | Ws_whole-plant | 10d-blue-light | GEO | Arabidopsis thaliana | GSE117797 | Download (14,500) |
| HY5 | Ws | Ws_whole-plant | 10d-red-light | GEO | Arabidopsis thaliana | GSE117797 | Download (9,536) |
| HY5 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (5,003) |
| ICE1 | HCT-116 | GEO | Homo sapiens | GSE47938 | Download (647) | ||
| ICE1 | HCT-116 | SHCTR | GEO | Homo sapiens | GSE47938 | Download (163) | |
| ICE1 | HCT-116 | SHICE1 | GEO | Homo sapiens | GSE47938 | Download (44) | |
| ICE2 | HCT-116 | GEO | Homo sapiens | GSE47938 | Download (2,228) | ||
| ID3 | K-562 | ENCODE | Homo sapiens | ENCSR005NMT | Download (49,419) | ||
| IDD4 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (991) |
| IKZF1 | BCR-ABL1 | GEO | Homo sapiens | GSE58825 | Download (7,935) | ||
| IKZF1 | BCR-ABL1 | LAX2 | GEO | Homo sapiens | GSE58825 | Download (22,316) | |
| IKZF1 | K-562 | ENCODE | Homo sapiens | ENCSR948VFL | Download (82,095) | ||
| IKZF1 | HSPC | GEO | Homo sapiens | GSE26014 | Download (1,682) | ||
| IKZF1 | GM12878 | ENCODE | Homo sapiens | ENCSR000EUJ | Download (1,006) | ||
| IKZF1 | GM12878 | ENCODE | Homo sapiens | ENCSR441VHN | Download (83,804) | ||
| IKZF1 | GM12878 | ENCODE | Homo sapiens | ENCSR874AFU | Download (86,020) | ||
| IKZF1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR278JQG | Download (3,536) | ||
| IKZF1 | K-562 | ENCODE | Homo sapiens | ENCSR395HWC | Download (73,796) | ||
| IKZF2 | GM12878 | ENCODE | Homo sapiens | ENCSR680UQE | Download (59,220) | ||
| IKZF2 | GM12878 | ENCODE | Homo sapiens | ENCSR822AHX | Download (45,108) | ||
| IKZF3 | HEK293 | ENCODE | Homo sapiens | ENCSR304AMN | Download (46,486) | ||
| IKZF5 | Hep-G2 | ENCODE | Homo sapiens | ENCSR210HBN | Download (84,001) | ||
| ILF3 | K-562 | ENCODE | Homo sapiens | ENCSR632TJQ | Download (2,618) | ||
| ILF3 | K-562 | GEO | Homo sapiens | GSE103215 | Download (52,842) | ||
| ILK | K-562 | ENCODE | Homo sapiens | ENCSR648INT | Download (2,605) | ||
| INO80 | Hep-G2 | GEO | Homo sapiens | GSE107730 | Download (21,974) | ||
| INSM2 | HEK293 | ENCODE | Homo sapiens | ENCSR382GSF | Download (21,647) | ||
| INTS11 | HL-60 | GEO | Homo sapiens | GSE106359 | Download (16,412) | ||
| INTS11 | HL-60 | PMA | GEO | Homo sapiens | GSE106359 | Download (12,979) | |
| INTS13 | monocyte | GEO | Homo sapiens | GSE106359 | Download (15,094) | ||
| INTS13 | HL-60 | GEO | Homo sapiens | GSE106359 | Download (17,622) | ||
| INTS13 | HL-60 | PMA | GEO | Homo sapiens | GSE106359 | Download (20,224) | |
| INTS13 | HL-60 | shEGR1 | GEO | Homo sapiens | GSE106359 | Download (10,268) | |
| IRF1 | monocyte | nopretreatment | GEO | Homo sapiens | GSE100381 | Download (5,212) | |
| IRF1 | monocyte | notreatment | GEO | Homo sapiens | GSE100381 | Download (15,743) | |
| IRF1 | K-562 | ENCODE | Homo sapiens | ENCSR000EGK | Download (1,944) | ||
| IRF1 | K-562 | ENCODE | Homo sapiens | ENCSR000EGL | Download (3,113) | ||
| IRF1 | K-562 | ENCODE | Homo sapiens | ENCSR000EGT | Download (9,751) | ||
| IRF1 | K-562 | ENCODE | Homo sapiens | ENCSR000EGU | Download (683) | ||
| IRF1 | K-562 | ENCODE | Homo sapiens | ENCSR854MCV | Download (17,154) | ||
| IRF1 | CD14 | GEO | Homo sapiens | GSE43036 | Download (366) | ||
| IRF1 | CD14 | LPS | GEO | Homo sapiens | GSE43036 | Download (11,054) | |
| IRF1 | HAEC | IL1b_4h | GEO | Homo sapiens | GSE89970 | Download (8,174) | |
| IRF1 | HAEC | TNFa_4h | GEO | Homo sapiens | GSE89970 | Download (4,708) | |
| IRF1 | PDAC | GEO | Homo sapiens | GSE64557 | Download (38,890) | ||
| IRF2 | CD34 | ADULT | GEO | Homo sapiens | GSE70660 | Download (4,921) | |
| IRF2 | K-562 | ENCODE | Homo sapiens | ENCSR376WCJ | Download (24,024) | ||
| IRF3 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZX | Download (463) | ||
| IRF3 | GM12878 | ENCODE | Homo sapiens | ENCSR408JQO | Download (3,687) | ||
| IRF3 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEJ | Download (582) | ||
| IRF3 | SK-N-SH | ENCODE | Homo sapiens | ENCSR422ZAO | Download (2,712) | ||
| IRF3 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EDF | Download (1,399) | ||
| IRF3 | HBTEC | E1Ainfected_FOXKb | GEO | Homo sapiens | GSE105037 | Download (146) | |
| IRF4 | GM12878 | ENCODE | Homo sapiens | ENCSR000BGY | Download (57,248) | ||
| IRF4 | NCI-H929 | GEO | Homo sapiens | GSE56857 | Download (40,496) | ||
| IRF4 | OCI-Ly3 | GEO | Homo sapiens | GSE56857 | Download (9,085) | ||
| IRF4 | OCI-Ly3 | SHCTR | GEO | Homo sapiens | GSE56857 | Download (8,295) | |
| IRF4 | OCI-Ly3 | SHSPIB | GEO | Homo sapiens | GSE56857 | Download (747) | |
| IRF4 | MM1-S | GEO | Homo sapiens | GSE49224 | Download (81,051) | ||
| IRF4 | KK-1 | GEO | Homo sapiens | GSE94732 | Download (5,072) | ||
| IRF5 | GM12878 | ENCODE | Homo sapiens | ENCSR976TBC | Download (4,695) | ||
| IRF9 | K-562 | ENCODE | Homo sapiens | ENCSR926KTP | Download (6,835) | ||
| ISL1 | SK-N-BE2-C | GEO | Homo sapiens | GSE94822 | Download (55,428) | ||
| JARID2 | AMIPS6 | GEO | Homo sapiens | GSE48516 | Download (12,580) | ||
| JARID2 | AMIPS8 | GEO | Homo sapiens | GSE48516 | Download (3,898) | ||
| JARID2 | EDOMIPS2 | GEO | Homo sapiens | GSE48516 | Download (5,289) | ||
| JARID2 | UTEIPS11 | GEO | Homo sapiens | GSE48516 | Download (5,849) | ||
| JARID2 | UTEIPS4 | GEO | Homo sapiens | GSE48516 | Download (3,467) | ||
| JARID2 | UTEIPS6 | GEO | Homo sapiens | GSE48516 | Download (14,009) | ||
| JARID2 | UTEIPS7 | GEO | Homo sapiens | GSE48516 | Download (6,904) | ||
| JARID2 | MRC-5 | IPS25_CTR | GEO | Homo sapiens | GSE48515 | Download (24,538) | |
| JARID2 | MRC-5 | IPS25_MEG3 | GEO | Homo sapiens | GSE48515 | Download (33,181) | |
| JARID2 | MRC-5 | IPS25 | GEO | Homo sapiens | GSE48516 | Download (3,613) | |
| JDP2 | Loucy | GEO | Homo sapiens | GSE115465 | Download (1,550) | ||
| JKD | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (226) |
| JMJD1C | NB4 | GEO | Homo sapiens | GSE63484 | Download (52,835) | ||
| JMJD1C | HL-60 | GEO | Homo sapiens | GSE63484 | Download (79,500) | ||
| JMJD1C | Kasumi-1 | GEO | Homo sapiens | GSE63484 | Download (45,574) | ||
| JMJD1C | THP-1 | GEO | Homo sapiens | GSE63484 | Download (40,486) | ||
| JMJD6 | HEK293T | GEO | Homo sapiens | GSE51633 | Download (4,618) | ||
| JMJD6 | HeLa | GEO | Homo sapiens | GSE51633 | Download (10,410) | ||
| JUB1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,894) |
| JUN | HAEC | GEO | Homo sapiens | GSE89970 | Download (110,958) | ||
| JUN | HAEC | IL1b_4h | GEO | Homo sapiens | GSE89970 | Download (69,962) | |
| JUN | HAEC | oxpapc_4h | GEO | Homo sapiens | GSE89970 | Download (27,003) | |
| JUN | HAEC | TNFa_4h | GEO | Homo sapiens | GSE89970 | Download (73,605) | |
| JUN | MCF-7 | ENCODE | Homo sapiens | ENCSR176EXN | Download (3,198) | ||
| JUN | MCF-7 | E2 | GEO | Homo sapiens | GSE102410 | Download (26,947) | |
| JUN | MCF-7 | Tamoxifen | GEO | Homo sapiens | GSE102410 | Download (32,989) | |
| JUN | MCF-7 | vehicle | GEO | Homo sapiens | GSE102410 | Download (8,321) | |
| JUN | WA01 | ENCODE | Homo sapiens | ENCSR000ECA | Download (2,177) | ||
| JUN | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EDG | Download (25,536) | ||
| JUN | endothelial | umbilical-vein | ENCODE | Homo sapiens | ENCSR000EFA | Download (42,258) | |
| JUN | A-549 | ENCODE | Homo sapiens | ENCSR996DUT | Download (4,262) | ||
| JUN | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEK | Download (24,518) | ||
| JUN | K-562 | ENCODE | Homo sapiens | ENCSR000EFS | Download (42,189) | ||
| JUN | K-562 | ENCODE | Homo sapiens | ENCSR000EGH | Download (5,466) | ||
| JUN | K-562 | ENCODE | Homo sapiens | ENCSR000EZT | Download (7,182) | ||
| JUN | K-562 | ENCODE | Homo sapiens | ENCSR000EZW | Download (5,116) | ||
| JUN | K-562 | ENCODE | Homo sapiens | ENCSR000EZX | Download (3,480) | ||
| JUN | K-562 | ENCODE | Homo sapiens | ENCSR000FAH | Download (31,429) | ||
| JUN | Calu-3 | GEO | Homo sapiens | GSE85401 | Download (4,578) | ||
| JUN | BT-549 | GEO | Homo sapiens | GSE46166 | Download (18,628) | ||
| JUN | BT-549 | GEO | Homo sapiens | GSE71976 | Download (4,592) | ||
| JUN | BT-549 | TNF | GEO | Homo sapiens | GSE71976 | Download (4,582) | |
| JUN | 786-O | GEO | Homo sapiens | GSE86092 | Download (72,544) | ||
| JUN | HUVEC-C | GEO | Homo sapiens | GSE109625 | Download (2,567) | ||
| JUN | HUVEC-C | VEGF_12h | GEO | Homo sapiens | GSE109625 | Download (3,270) | |
| JUN | HUVEC-C | VEGF_1h | GEO | Homo sapiens | GSE109625 | Download (4,168) | |
| JUN | HUVEC-C | VEGF_4h | GEO | Homo sapiens | GSE109625 | Download (614) | |
| JUN | MCF10A-Er-Src | EtOH | GEO | Homo sapiens | GSE115597 | Download (61,103) | |
| JUN | MCF10A-Er-Src | TAM | GEO | Homo sapiens | GSE115597 | Download (105,004) | |
| JUN | MDA-MB-231 | GEO | Homo sapiens | GSE66081 | Download (101,251) | ||
| JUNB | keratinocyte | neonatal | GEO | Homo sapiens | GSE63080 | Download (200) | |
| JUNB | MCF10A-Er-Src | EtOH | GEO | Homo sapiens | GSE115597 | Download (63,382) | |
| JUNB | MCF10A-Er-Src | TAM | GEO | Homo sapiens | GSE115597 | Download (87,033) | |
| JUNB | K-562 | ENCODE | Homo sapiens | ENCSR795IYP | Download (5,655) | ||
| JUNB | HAEC | GEO | Homo sapiens | GSE89970 | Download (50,629) | ||
| JUNB | GM12878 | ENCODE | Homo sapiens | ENCSR897MMC | Download (48,667) | ||
| JUNB | hESC | ENCODE | Homo sapiens | ENCSR917MAH | Download (602) | ||
| JUNB | K-562 | ENCODE | Homo sapiens | ENCSR000DJY | Download (40,145) | ||
| JUND | GM12878 | ENCODE | Homo sapiens | ENCSR000DYS | Download (5,365) | ||
| JUND | GM12878 | ENCODE | Homo sapiens | ENCSR000EYV | Download (5,216) | ||
| JUND | liver | ENCODE | Homo sapiens | ENCSR196HGZ | Download (30,021) | ||
| JUND | liver | ENCODE | Homo sapiens | ENCSR837GTK | Download (45,495) | ||
| JUND | A-549 | ENCODE | Homo sapiens | ENCSR000BRF | Download (37,116) | ||
| JUND | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BGK | Download (52,210) | ||
| JUND | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEI | Download (92,237) | ||
| JUND | K-562 | ENCODE | Homo sapiens | ENCSR000DJX | Download (81,806) | ||
| JUND | K-562 | ENCODE | Homo sapiens | ENCSR000EGN | Download (77,785) | ||
| JUND | MCF-7 | ENCODE | Homo sapiens | ENCSR000BSU | Download (16,461) | ||
| JUND | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BSK | Download (53,101) | ||
| JUND | SK-N-SH | ENCODE | Homo sapiens | ENCSR000EIB | Download (35,280) | ||
| JUND | T-47D | ENCODE | Homo sapiens | ENCSR000BVO | Download (8,998) | ||
| JUND | WA01 | ENCODE | Homo sapiens | ENCSR000BKP | Download (27,039) | ||
| JUND | WA01 | ENCODE | Homo sapiens | ENCSR000EBZ | Download (24,223) | ||
| JUND | HCT-116 | ENCODE | Homo sapiens | ENCSR000BSA | Download (25,404) | ||
| JUND | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EDH | Download (53,005) | ||
| JUND | GP5D | GEO | Homo sapiens | GSE51234 | Download (6,347) | ||
| JUND | GP5D | SIRAD21 | GEO | Homo sapiens | GSE51234 | Download (15,553) | |
| JUND | HFOB | DIFF | GEO | Homo sapiens | GSE82295 | Download (1,189) | |
| JUND | HT29 | DSMO | GEO | Homo sapiens | GSE77039 | Download (17,881) | |
| JUND | Calu-3 | GEO | Homo sapiens | GSE85401 | Download (5,861) | ||
| JUND | PC-3 | GEO | Homo sapiens | GSE29808 | Download (6,187) | ||
| KAN1 | Col-0 | Col-0_seedling | 10d-rep1 | GEO | Arabidopsis thaliana | GSE48081 | Download (13,845) |
| KAN2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (165) |
| KAT2A | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECR | Download (83) | ||
| KAT2B | A-549 | ENCODE | Homo sapiens | ENCSR356WVQ | Download (2,173) | ||
| KAT2B | Hep-G2 | ENCODE | Homo sapiens | ENCSR620YNB | Download (2,781) | ||
| KAT2B | K-562 | ENCODE | Homo sapiens | ENCSR000ATZ | Download (1,000) | ||
| KAT8 | Hep-G2 | ENCODE | Homo sapiens | ENCSR954KIC | Download (84,398) | ||
| KAT8 | K-562 | ENCODE | Homo sapiens | ENCSR086FZL | Download (611) | ||
| KDM1A | A-549 | ENCODE | Homo sapiens | ENCSR639GWS | Download (11,177) | ||
| KDM1A | Hep-G2 | ENCODE | Homo sapiens | ENCSR115BLD | Download (117,928) | ||
| KDM1A | K-562 | ENCODE | Homo sapiens | ENCSR000ATX | Download (15,393) | ||
| KDM1A | K-562 | ENCODE | Homo sapiens | ENCSR360HRA | Download (20,970) | ||
| KDM1A | K-562 | ENCODE | Homo sapiens | ENCSR908CMW | Download (34,794) | ||
| KDM1A | WA01 | ENCODE | Homo sapiens | ENCSR418RRF | Download (97,441) | ||
| KDM1A | HeLa | GEO | Homo sapiens | GSE45441 | Download (3,299) | ||
| KDM1A | T-47D-MTVL | GEO | Homo sapiens | GSE64467 | Download (1,736) | ||
| KDM1A | keratinocyte | diff | GEO | Homo sapiens | GSE57702 | Download (20,908) | |
| KDM1A | SKNO1 | DMSO | GEO | Homo sapiens | GSE71739 | Download (4,962) | |
| KDM1A | SKNO1 | GSK690 | GEO | Homo sapiens | GSE71739 | Download (27,744) | |
| KDM1A | SKNO1 | RN1 | GEO | Homo sapiens | GSE71739 | Download (26,643) | |
| KDM1A | Kasumi-1 | DMSO | GEO | Homo sapiens | GSE71739 | Download (38,193) | |
| KDM1A | Kasumi-1 | GSK690 | GEO | Homo sapiens | GSE71739 | Download (32,916) | |
| KDM1A | Kasumi-1 | RN1 | GEO | Homo sapiens | GSE71739 | Download (39,947) | |
| KDM1A | NCI-H526 | DMSO | GEO | Homo sapiens | GSE66297 | Download (35,002) | |
| KDM1A | SH-SY5Y | B0_SHLSD18A | GEO | Homo sapiens | GSE58258 | Download (139) | |
| KDM1A | SH-SY5Y | B3_SHCTR | GEO | Homo sapiens | GSE58258 | Download (949) | |
| KDM1A | SH-SY5Y | B3_SHLSD18A | GEO | Homo sapiens | GSE58258 | Download (102) | |
| KDM1A | SH-SY5Y | B0 | GEO | Homo sapiens | GSE58258 | Download (17,385) | |
| KDM1A | SH-SY5Y | B0_SHLSD18A | GEO | Homo sapiens | GSE58258 | Download (12,219) | |
| KDM1A | SH-SY5Y | B3 | GEO | Homo sapiens | GSE58258 | Download (44,347) | |
| KDM1B | Hep-G2 | GEO | Homo sapiens | GSE59694 | Download (2) | ||
| KDM3A | Hep-G2 | ENCODE | Homo sapiens | ENCSR387JKT | Download (56,988) | ||
| KDM4A | WA01 | ENCODE | Homo sapiens | ENCSR000AVC | Download (37,541) | ||
| KDM4B | K-562 | ENCODE | Homo sapiens | ENCSR642VZY | Download (23,923) | ||
| KDM4C | KYSE150 | GEO | Homo sapiens | GSE53938 | Download (40,302) | ||
| KDM5A | T-47D | DMSO | GEO | Homo sapiens | GSE80593 | Download (6,679) | |
| KDM5A | T-47D | MK2206 | GEO | Homo sapiens | GSE80593 | Download (8,340) | |
| KDM5A | WA01 | ENCODE | Homo sapiens | ENCSR000AQL | Download (2,912) | ||
| KDM5A | WA01 | ENCODE | Homo sapiens | ENCSR160ZLP | Download (3,856) | ||
| KDM5A | A-549 | ENCODE | Homo sapiens | ENCSR933MHJ | Download (5,282) | ||
| KDM5B | K-562 | ENCODE | Homo sapiens | ENCSR000AQA | Download (32,058) | ||
| KDM5B | WA01 | ENCODE | Homo sapiens | ENCSR000AUR | Download (1,826) | ||
| KDM5B | HCC2157 | GEO | Homo sapiens | GSE46055 | Download (59,819) | ||
| KDM5B | SUM159 | GEO | Homo sapiens | GSE46055 | Download (46,154) | ||
| KDM5B | SUM185 | GEO | Homo sapiens | GSE46055 | Download (16,029) | ||
| KDM5B | SUM185 | SHCTCF | GEO | Homo sapiens | GSE46055 | Download (56,311) | |
| KDM5B | MCF-7 | GEO | Homo sapiens | GSE46055 | Download (21,541) | ||
| KDM5B | MDA-MB-231 | GEO | Homo sapiens | GSE46055 | Download (105,734) | ||
| KDM5B | T-47D | GEO | Homo sapiens | GSE46055 | Download (54,083) | ||
| KDM6B | CUTLL1 | GEO | Homo sapiens | GSE56694 | Download (37,674) | ||
| KLF1 | erythroid | R3R4 | GEO | Homo sapiens | GSE43625 | Download (34,995) | |
| KLF1 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (25,216) | ||
| KLF1 | HEK293 | ENCODE | Homo sapiens | ENCSR859BMR | Download (48,308) | ||
| KLF1 | K-562 | ENCODE | Homo sapiens | ENCSR550HCT | Download (9,963) | ||
| KLF1 | HUDEP-2 | S2 | GEO | Homo sapiens | GSE97671 | Download (8,490) | |
| KLF1 | HUDEP-2 | S4 | GEO | Homo sapiens | GSE97671 | Download (10,992) | |
| KLF10 | Hep-G2 | GEO | Homo sapiens | GSE97661 | Download (16,635) | ||
| KLF10 | MCF-7 | GEO | Homo sapiens | GSE97661 | Download (4,453) | ||
| KLF10 | HEK293 | ENCODE | Homo sapiens | ENCSR006GAQ | Download (23,936) | ||
| KLF12 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (10,281) | ||
| KLF13 | K-562 | ENCODE | Homo sapiens | ENCSR608HVP | Download (16,109) | ||
| KLF13 | HEK293 | ENCODE | Homo sapiens | ENCSR946ZLI | Download (10,211) | ||
| KLF14 | HEK293 | ENCODE | Homo sapiens | ENCSR780ESQ | Download (14,310) | ||
| KLF15 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (8,981) | ||
| KLF16 | K-562 | ENCODE | Homo sapiens | ENCSR760UVO | Download (21,477) | ||
| KLF16 | HEK293 | ENCODE | Homo sapiens | ENCSR397DQC | Download (22,375) | ||
| KLF17 | HEK293 | ENCODE | Homo sapiens | ENCSR065WUF | Download (46,831) | ||
| KLF3 | HEK293 | GEO | Homo sapiens | GSE69739 | Download (2,899) | ||
| KLF4 | BJ | INDUCED | GEO | Homo sapiens | GSE36570 | Download (55,036) | |
| KLF4 | WA09 | GEO | Homo sapiens | GSE105028 | Download (2,012) | ||
| KLF4 | WA09 | heat-shock | GEO | Homo sapiens | GSE105028 | Download (690) | |
| KLF4 | hESC | GEO | Homo sapiens | GSE17917 | Download (15) | ||
| KLF4 | keratinocyte | diff | GEO | Homo sapiens | GSE57702 | Download (41,294) | |
| KLF4 | PDAC | GEO | Homo sapiens | GSE64557 | Download (44,924) | ||
| KLF4 | MCF-7 | GEO | Homo sapiens | GSE41561 | Download (7,711) | ||
| KLF4 | hiPSC | GEO | Homo sapiens | GSE56567 | Download (8,040) | ||
| KLF4 | OSK | GEO | Homo sapiens | GSE81899 | Download (218) | ||
| KLF5 | BICR | -31 | GEO | Homo sapiens | GSE88976 | Download (610) | |
| KLF5 | GM12878 | ENCODE | Homo sapiens | ENCSR974OFJ | Download (11,406) | ||
| KLF5 | AGS | GEO | Homo sapiens | GSE51705 | Download (775) | ||
| KLF5 | GP5D | GEO | Homo sapiens | GSE51234 | Download (15,270) | ||
| KLF5 | GP5D | SIRAD21 | GEO | Homo sapiens | GSE51234 | Download (377) | |
| KLF5 | HCC95 | GEO | Homo sapiens | GSE88976 | Download (10,804) | ||
| KLF5 | HCC95 | E419Q | GEO | Homo sapiens | GSE88976 | Download (4,329) | |
| KLF5 | HEK293 | GEO | Homo sapiens | GSE88976 | Download (15,094) | ||
| KLF5 | HEK293 | D418N | GEO | Homo sapiens | GSE88976 | Download (19,525) | |
| KLF5 | HEK293 | E419K | GEO | Homo sapiens | GSE88976 | Download (14,314) | |
| KLF5 | HEK293 | E419Q | GEO | Homo sapiens | GSE88976 | Download (20,539) | |
| KLF5 | KATOIII | GEO | Homo sapiens | GSE51705 | Download (27,570) | ||
| KLF5 | LoVo | PHASEM | GEO | Homo sapiens | GSE51290 | Download (356) | |
| KLF5 | LoVo | PHASES | GEO | Homo sapiens | GSE51290 | Download (2,665) | |
| KLF5 | YCC-3 | GEO | Homo sapiens | GSE51705 | Download (2,475) | ||
| KLF6 | PDAC | GEO | Homo sapiens | GSE64557 | Download (48,221) | ||
| KLF6 | Hep-G2 | ENCODE | Homo sapiens | ENCSR757EKM | Download (62,548) | ||
| KLF7 | HEK293 | ENCODE | Homo sapiens | ENCSR604VWJ | Download (11,025) | ||
| KLF7 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (582) | ||
| KLF8 | HEK293 | ENCODE | Homo sapiens | ENCSR635NOQ | Download (38,188) | ||
| KLF9 | HEK293 | ENCODE | Homo sapiens | ENCSR076EZB | Download (25,821) | ||
| KLF9 | MCF-7 | ENCODE | Homo sapiens | ENCSR125ZYC | Download (11,710) | ||
| KLF9 | GBM1A | GEO | Homo sapiens | GSE62211 | Download (79,837) | ||
| KMT2A | CD34 | GEO | Homo sapiens | GSE54344 | Download (5) | ||
| KMT2A | SEM | GEO | Homo sapiens | GSE83671 | Download (27,596) | ||
| KMT2A | CCRF-CEM | GEO | Homo sapiens | GSE83671 | Download (28,138) | ||
| KMT2A | KOPN-8 | GEO | Homo sapiens | GSE83671 | Download (23,875) | ||
| KMT2A | MV4-11 | GEO | Homo sapiens | GSE83671 | Download (23,248) | ||
| KMT2A | RCH-ACV | GEO | Homo sapiens | GSE83671 | Download (52,056) | ||
| KMT2A | THP-1 | GEO | Homo sapiens | GSE83671 | Download (14,040) | ||
| KMT2A | RS4-11 | GEO | Homo sapiens | GSE38403 | Download (9,818) | ||
| KMT2A | blood | cord | GEO | Homo sapiens | GSE83671 | Download (48,796) | |
| KMT2A | bone-marrow | fetal | GEO | Homo sapiens | GSE83671 | Download (87,387) | |
| KMT2A | L826 | GEO | Homo sapiens | GSE83671 | Download (41,659) | ||
| KMT2A | SHI-1 | GEO | Homo sapiens | GSE95511 | Download (11,964) | ||
| KMT2B | AML | GEO | Homo sapiens | GSE112074 | Download (32,673) | ||
| KMT2B | AML | OG86 | GEO | Homo sapiens | GSE112074 | Download (53,937) | |
| KMT2B | B-cell | GERMINAL_CENTER_TONSIL | GEO | Homo sapiens | GSE67494 | Download (3,090) | |
| KMT2D | MCF-7 | GEO | Homo sapiens | GSE85317 | Download (6,231) | ||
| KUA1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,103) |
| L3MBTL2 | K-562 | GEO | Homo sapiens | GSE28162 | Download (2,909) | ||
| L3MBTL2 | HEK293T | ENCODE | Homo sapiens | ENCSR862PNL | Download (17,485) | ||
| L3MBTL2 | K-562 | ENCODE | Homo sapiens | ENCSR530XQI | Download (56,038) | ||
| L3MBTL4 | Hep-G2 | GEO | Homo sapiens | GSE97661 | Download (12,219) | ||
| LBD13 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (124) |
| LBD13 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (3,211) |
| LBD16 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (3,695) |
| LBD18 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,714) |
| LBD18 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (16,315) |
| LBD19 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,050) |
| LBD23 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (130) |
| LCORL | Hep-G2 | ENCODE | Homo sapiens | ENCSR950NAZ | Download (9,495) | ||
| LDB1 | HEP | GEO | Homo sapiens | GSE52637 | Download (39,521) | ||
| LDL1 | Col-0 | Col-0_seedling | 14d | GEO | Arabidopsis thaliana | GSE118025 | Download (5,626) |
| LEF1 | hESC | GEO | Homo sapiens | GSE64758 | Download (442) | ||
| LEF1 | hESC | WNT3A | GEO | Homo sapiens | GSE64758 | Download (1,010) | |
| LEF1 | K-562 | ENCODE | Homo sapiens | ENCSR832OGB | Download (1,010) | ||
| LEF1 | HEK293T | ENCODE | Homo sapiens | ENCSR240XWM | Download (288) | ||
| LEF1 | K-562 | ENCODE | Homo sapiens | ENCSR343ELW | Download (20,867) | ||
| LEO1 | SEM | GEO | Homo sapiens | GSE83671 | Download (93,224) | ||
| LEP | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (479) |
| LFY | Col-0 | Col-0_seedling | 15d | GEO | Arabidopsis thaliana | GSE24568 | Download (5,770) |
| LFY | Col-0 | Col-0_seedling | 15d-LFY | GEO | Arabidopsis thaliana | GSE64245 | Download (10,429) |
| LFY-TERE | Col-0 | Col-0_seedling | 15d-LFY-TERE | GEO | Arabidopsis thaliana | GSE64245 | Download (3,929) |
| LHX2 | retina | pigment | GEO | Homo sapiens | GSE60024 | Download (22,645) | |
| LHY | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,228) |
| LHY | Ws | Ws_seedling | 2w-mutant | GEO | Arabidopsis thaliana | GSE52175 | Download (18,257) |
| LHY | Ws | Ws_seedling | 2w-wt | GEO | Arabidopsis thaliana | GSE52175 | Download (5,505) |
| LMO1 | Jurkat | GEO | Homo sapiens | GSE94391 | Download (43,562) | ||
| LMO2 | CCRF-CEM | GEO | Homo sapiens | GSE33850 | Download (64,084) | ||
| LMO2 | Kasumi-1 | GEO | Homo sapiens | GSE43834 | Download (6,524) | ||
| LMO2 | Kasumi-1 | SICTR | GEO | Homo sapiens | GSE60130 | Download (33,021) | |
| LMO2 | Kasumi-1 | SIRUNX1ETO | GEO | Homo sapiens | GSE60130 | Download (23,373) | |
| LMO2 | TSU-1621MT | GEO | Homo sapiens | GSE60477 | Download (32,044) | ||
| LOB | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (784) |
| LRWD1 | HeLa | GEO | Homo sapiens | GSE20303 | Download (3) | ||
| LYL1 | NB4 | GEO | Homo sapiens | GSE63484 | Download (4,957) | ||
| LYL1 | Kasumi-1 | GEO | Homo sapiens | GSE63484 | Download (10,454) | ||
| LYL1 | THP-1 | GEO | Homo sapiens | GSE63484 | Download (5,635) | ||
| LYL1 | TSU-1621MT | GEO | Homo sapiens | GSE60477 | Download (12,663) | ||
| MAF | keratinocyte | epidermal_PROLIF | GEO | Homo sapiens | GSE52953 | Download (3,726) | |
| MAF | CD4 | TH1 | GEO | Homo sapiens | GSE72266 | Download (59,967) | |
| MAF | CD4 | TH2 | GEO | Homo sapiens | GSE72266 | Download (31,386) | |
| MAF | lymphocyte | Th17_IL10+_Day5 | GEO | Homo sapiens | GSE101389 | Download (12,353) | |
| MAF | monocyte | GEO | Homo sapiens | GSE98367 | Download (2,017) | ||
| MAF | monocyte | INFg | GEO | Homo sapiens | GSE98367 | Download (1,927) | |
| MAF1 | THP-1 | macrophage_PMA | GEO | Homo sapiens | GSE96800 | Download (21,515) | |
| MAF1 | THP-1 | monocytes | GEO | Homo sapiens | GSE96800 | Download (3,616) | |
| MAF1 | Col-0 | Col-0_seedling | 10d-FLMbeta | GEO | Arabidopsis thaliana | GSE48082 | Download (290) |
| MAF1 | Col-0 | Col-0_seedling | 10d-genomicFLM | GEO | Arabidopsis thaliana | GSE48082 | Download (4,529) |
| MAFB | keratinocyte | epidermal_PROLIF | GEO | Homo sapiens | GSE52953 | Download (5,653) | |
| MAFB | islet | ENA | Homo sapiens | ERP004003 | Download (53,287) | ||
| MAFF | GM12878 | ENCODE | Homo sapiens | ENCSR237YZZ | Download (6,053) | ||
| MAFF | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEC | Download (82,791) | ||
| MAFF | K-562 | ENCODE | Homo sapiens | ENCSR000EGI | Download (29,370) | ||
| MAFF | HeLa-S3 | ENCODE | Homo sapiens | ENCSR140DSL | Download (69,004) | ||
| MAFG | K-562 | ENCODE | Homo sapiens | ENCSR818DQV | Download (56,561) | ||
| MAFK | MCF-7 | ENCODE | Homo sapiens | ENCSR555PBN | Download (10,392) | ||
| MAFK | WA01 | ENCODE | Homo sapiens | ENCSR000EBS | Download (15,210) | ||
| MAFK | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECK | Download (19,826) | ||
| MAFK | GM12878 | ENCODE | Homo sapiens | ENCSR000DYV | Download (1,712) | ||
| MAFK | A-549 | ENCODE | Homo sapiens | ENCSR541WQI | Download (90,712) | ||
| MAFK | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EDZ | Download (65,589) | ||
| MAFK | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEB | Download (94,493) | ||
| MAFK | IMR-90 | ENCODE | Homo sapiens | ENCSR000EFH | Download (93,646) | ||
| MAFK | K-562 | ENCODE | Homo sapiens | ENCSR000EGX | Download (23,798) | ||
| MAFK | OCI-Ly7 | GEO | Homo sapiens | GSE47784 | Download (21,288) | ||
| MAML3 | SK-N-SH | GEO | Homo sapiens | GSE69119 | Download (8,977) | ||
| MAML3 | SK-N-SH | RA | GEO | Homo sapiens | GSE69119 | Download (3,606) | |
| MAX | MDA-MB-468 | GEO | Homo sapiens | GSE81381 | Download (31,789) | ||
| MAX | MKL-1 | GEO | Homo sapiens | GSE69877 | Download (110,398) | ||
| MAX | NCI-H2171 | GEO | Homo sapiens | GSE36354 | Download (79,100) | ||
| MAX | NCI-H2171 | GEO | Homo sapiens | GSE41105 | Download (80,226) | ||
| MAX | P493-6 | GEO | Homo sapiens | GSE36354 | Download (49,269) | ||
| MAX | P493-6 | CMYC_1H | GEO | Homo sapiens | GSE36354 | Download (38,471) | |
| MAX | P493-6 | CMYC_24H | GEO | Homo sapiens | GSE36354 | Download (18,841) | |
| MAX | P493-6 | GEO | Homo sapiens | GSE42262 | Download (27,954) | ||
| MAX | MCF-7 | ENCODE | Homo sapiens | ENCSR000BUL | Download (67,050) | ||
| MAX | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BVD | Download (62,829) | ||
| MAX | WA01 | ENCODE | Homo sapiens | ENCSR000BSJ | Download (108,406) | ||
| MAX | WA01 | ENCODE | Homo sapiens | ENCSR000EUP | Download (24,844) | ||
| MAX | HCT-116 | ENCODE | Homo sapiens | ENCSR000BSH | Download (25,649) | ||
| MAX | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECN | Download (45,371) | ||
| MAX | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EZF | Download (56,449) | ||
| MAX | H128 | GEO | Homo sapiens | GSE41105 | Download (26,463) | ||
| MAX | U-87MG | GEO | Homo sapiens | GSE36354 | Download (98,698) | ||
| MAX | MM1-S | JQ1 | GEO | Homo sapiens | GSE42161 | Download (298) | |
| MAX | endothelial | umbilical-vein | ENCODE | Homo sapiens | ENCSR000EEZ | Download (25,524) | |
| MAX | GM12878 | ENCODE | Homo sapiens | ENCSR000DZF | Download (3,694) | ||
| MAX | Ishikawa | ENCODE | Homo sapiens | ENCSR000BTY | Download (53,272) | ||
| MAX | liver | ENCODE | Homo sapiens | ENCSR521IID | Download (34,559) | ||
| MAX | liver | ENCODE | Homo sapiens | ENCSR847DIT | Download (43,893) | ||
| MAX | NB4 | ENCODE | Homo sapiens | ENCSR000EHS | Download (13,556) | ||
| MAX | A-549 | ENCODE | Homo sapiens | ENCSR000BTJ | Download (46,199) | ||
| MAX | A-549 | ENCODE | Homo sapiens | ENCSR000DYG | Download (15,602) | ||
| MAX | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BTM | Download (58,767) | ||
| MAX | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EDS | Download (32,215) | ||
| MAX | K-562 | ENCODE | Homo sapiens | ENCSR000BLP | Download (123,763) | ||
| MAX | K-562 | ENCODE | Homo sapiens | ENCSR000EFV | Download (75,998) | ||
| MAX | K-562 | ENCODE | Homo sapiens | ENCSR000FAE | Download (9,793) | ||
| MAZ | GM12878 | ENCODE | Homo sapiens | ENCSR000DZA | Download (7,685) | ||
| MAZ | HEK293 | ENCODE | Homo sapiens | ENCSR290SSQ | Download (37,937) | ||
| MAZ | A-549 | ENCODE | Homo sapiens | ENCSR636YLV | Download (10,164) | ||
| MAZ | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EDN | Download (24,607) | ||
| MAZ | IMR-90 | ENCODE | Homo sapiens | ENCSR000EFF | Download (56,163) | ||
| MAZ | K-562 | ENCODE | Homo sapiens | ENCSR000EFX | Download (60,085) | ||
| MAZ | MCF-7 | ENCODE | Homo sapiens | ENCSR288IJC | Download (17,846) | ||
| MAZ | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECL | Download (25,018) | ||
| MAZ | HEK293 | GEO | Homo sapiens | GSE76494 | Download (44,410) | ||
| MAZ | T778 | GEO | Homo sapiens | GSE57499 | Download (302,424) | ||
| MBD1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR260UJI | Download (20,908) | ||
| MBD1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR396QWK | Download (14,643) | ||
| MBD2 | K-562 | ENCODE | Homo sapiens | ENCSR221GAN | Download (18,485) | ||
| MBD2 | HMLER | GEO | Homo sapiens | GSE63233 | Download (441) | ||
| MBD2 | HMEC-1 | GEO | Homo sapiens | GSE63233 | Download (969) | ||
| MBD2 | MCF-7 | GEO | Homo sapiens | GSE54693 | Download (241) | ||
| MBD2 | MCF-7 | ENCODE | Homo sapiens | ENCSR940MHE | Download (23,663) | ||
| MBD2 | HeLa | GEO | Homo sapiens | GSE41006 | Download (5,943) | ||
| MBD3 | HeLa | GEO | Homo sapiens | GSE41006 | Download (37) | ||
| MBD3 | MCF-7 | GEO | Homo sapiens | GSE44737 | Download (19,778) | ||
| MBD4 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BQW | Download (4,447) | ||
| MBD7 | Col-0 | Col-0_seedling | 14d-wt | GEO | Arabidopsis thaliana | GSE58789 | Download (231) |
| MCM2 | K-562 | ENCODE | Homo sapiens | ENCSR552PSV | Download (1,056) | ||
| MCM3 | K-562 | ENCODE | Homo sapiens | ENCSR990AZC | Download (22,807) | ||
| MCM5 | K-562 | ENCODE | Homo sapiens | ENCSR628APV | Download (5,590) | ||
| MCM5 | K-562 | ENCODE | Homo sapiens | ENCSR079WHK | Download (179) | ||
| MCM7 | K-562 | ENCODE | Homo sapiens | ENCSR038RGL | Download (1,142) | ||
| MCM7 | K-562 | ENCODE | Homo sapiens | ENCSR068VRA | Download (197) | ||
| MCM7 | K-562 | ENCODE | Homo sapiens | ENCSR542WJU | Download (266) | ||
| MCRS1 | Hep-G2 | GEO | Homo sapiens | GSE107730 | Download (20,630) | ||
| MDM2 | H1299 | GEO | Homo sapiens | GSE64439 | Download (1) | ||
| MECOM | SKH1 | GEO | Homo sapiens | GSE102697 | Download (15,220) | ||
| MECOM | SKH1 | CEBPA-ER | GEO | Homo sapiens | GSE102697 | Download (28,335) | |
| MECOM | SKH1 | CEBPA-ER_E2 | GEO | Homo sapiens | GSE102697 | Download (24,981) | |
| MECOM | SKH1 | E2 | GEO | Homo sapiens | GSE102697 | Download (19,393) | |
| MED | SEM | GEO | Homo sapiens | GSE83671 | Download (31,551) | ||
| MED1 | MM1-S | DMSO | GEO | Homo sapiens | GSE36354 | Download (885) | |
| MED1 | U-87MG | GEO | Homo sapiens | GSE36354 | Download (59,424) | ||
| MED1 | MM1-S | JQ1 | GEO | Homo sapiens | GSE42161 | Download (35,446) | |
| MED1 | MM1-S | JQ1_500NM | GEO | Homo sapiens | GSE42355 | Download (43,642) | |
| MED1 | MM1-S | BIORU | GEO | Homo sapiens | GSE45984 | Download (2,449) | |
| MED1 | MM1-S | JQ1_5000NM | GEO | Homo sapiens | GSE49224 | Download (37,607) | |
| MED1 | MM1-S | JQ1_50NM | GEO | Homo sapiens | GSE49224 | Download (53,616) | |
| MED1 | MM1-S | JQ1_5NM | GEO | Homo sapiens | GSE49224 | Download (51,987) | |
| MED1 | cardiomyocyte | GEO | Homo sapiens | GSE85628 | Download (2,143) | ||
| MED1 | cardiomyocyte | 1 | GEO | Homo sapiens | GSE85628 | Download (2,148) | |
| MED1 | cardiomyocyte | 5 | GEO | Homo sapiens | GSE85628 | Download (18,647) | |
| MED1 | cardiomyocyte | 7 | GEO | Homo sapiens | GSE85628 | Download (7,834) | |
| MED1 | G296S | GEO | Homo sapiens | GSE85628 | Download (14,809) | ||
| MED1 | G296S | 2 | GEO | Homo sapiens | GSE85628 | Download (14,814) | |
| MED1 | G296S | 4 | GEO | Homo sapiens | GSE85628 | Download (21,341) | |
| MED1 | dopaminergic-neuron | Dopamine_neurons | GEO | Homo sapiens | GSE93905 | Download (11,131) | |
| MED1 | K-562 | GEO | Homo sapiens | GSE97661 | Download (3,440) | ||
| MED1 | MDA-MB-231 | GEO | Homo sapiens | GSE95121 | Download (728) | ||
| MED1 | MDA-MB-231 | 45min | GEO | Homo sapiens | GSE95121 | Download (562) | |
| MED1 | MDA-MB-231 | LQ | GEO | Homo sapiens | GSE95121 | Download (5,340) | |
| MED1 | MDA-MB-231 | LQ_45min | GEO | Homo sapiens | GSE95121 | Download (4,036) | |
| MED1 | hMSC-TERT4 | 4h | GEO | Homo sapiens | GSE104537 | Download (67,969) | |
| MED1 | hMSC-TERT4 | D0 | GEO | Homo sapiens | GSE104537 | Download (17,740) | |
| MED1 | hMSC-TERT4 | D1 | GEO | Homo sapiens | GSE104537 | Download (5,854) | |
| MED1 | hMSC-TERT4 | D14 | GEO | Homo sapiens | GSE104537 | Download (36,804) | |
| MED1 | hMSC-TERT4 | D3 | GEO | Homo sapiens | GSE104537 | Download (50,414) | |
| MED1 | hMSC-TERT4 | D7 | GEO | Homo sapiens | GSE104537 | Download (44,994) | |
| MED1 | hESC | PRIMED | GEO | Homo sapiens | GSE69646 | Download (11,745) | |
| MED1 | Jurkat | GEO | Homo sapiens | GSE59657 | Download (13,699) | ||
| MED1 | LS180 | GEO | Homo sapiens | GSE39277 | Download (2,720) | ||
| MED1 | LS180 | 125 | GEO | Homo sapiens | GSE39277 | Download (1,550) | |
| MED1 | myoblast | GEO | Homo sapiens | GSE60026 | Download (11,999) | ||
| MED1 | SGBS | GEO | Homo sapiens | GSE64233 | Download (44,633) | ||
| MED1 | SGBS | TNF | GEO | Homo sapiens | GSE64233 | Download (28,864) | |
| MED1 | A-549 | GEO | Homo sapiens | GSE76893 | Download (13,628) | ||
| MED1 | Hep-G2 | GEO | Homo sapiens | GSE76893 | Download (18,242) | ||
| MED1 | MCF-7 | GEO | Homo sapiens | GSE60270 | Download (35) | ||
| MED1 | MCF-7 | E2 | GEO | Homo sapiens | GSE60270 | Download (6,678) | |
| MED1 | MCF-7 | SHCTR | GEO | Homo sapiens | GSE60270 | Download (23,389) | |
| MED1 | MCF-7 | SHCTR_E2 | GEO | Homo sapiens | GSE60270 | Download (17,747) | |
| MED1 | MCF-7 | SHRARS | GEO | Homo sapiens | GSE60270 | Download (47,398) | |
| MED1 | MCF-7 | SHRARS_E2 | GEO | Homo sapiens | GSE60270 | Download (36,428) | |
| MED1 | MCF-7 | GEO | Homo sapiens | GSE76893 | Download (4,736) | ||
| MED1 | MOLM-14 | GEO | Homo sapiens | GSE65138 | Download (18,935) | ||
| MED1 | NCI-H2171 | GEO | Homo sapiens | GSE36354 | Download (14,506) | ||
| MED1 | OCI-Ly1 | GEO | Homo sapiens | GSE53601 | Download (3,391) | ||
| MED1 | P493-6 | GEO | Homo sapiens | GSE36354 | Download (12,607) | ||
| MED1 | P493-6 | CMYC_1H | GEO | Homo sapiens | GSE36354 | Download (2,484) | |
| MED1 | P493-6 | CMYC_24H | GEO | Homo sapiens | GSE36354 | Download (1,066) | |
| MED1 | RH4 | GEO | Homo sapiens | GSE83726 | Download (28,096) | ||
| MED1 | SUM159PT | 100nMtrametinib_24h | GEO | Homo sapiens | GSE87418 | Download (21,656) | |
| MED1 | SUM159PT | 100nMtrametinib300nMJQ1_24h | GEO | Homo sapiens | GSE87418 | Download (12,497) | |
| MED1 | SUM159PT | 300nMJQ1_24h | GEO | Homo sapiens | GSE87418 | Download (23,966) | |
| MED1 | SUM159PT | DMSO_24h | GEO | Homo sapiens | GSE87418 | Download (23,416) | |
| MED1 | CD4 | TH1_BAY | GEO | Homo sapiens | GSE62482 | Download (8,984) | |
| MED1 | CD4 | TH1_DMSO | GEO | Homo sapiens | GSE62482 | Download (10,364) | |
| MED1 | GM12878 | GEO | Homo sapiens | GSE93080 | Download (41,604) | ||
| MED12 | MCF-7 | siCTL_E2 | GEO | Homo sapiens | GSE101559 | Download (1,473) | |
| MED26 | U2OS | SHCTR | GEO | Homo sapiens | GSE73742 | Download (23,792) | |
| MED26 | U2OS | SHEP400 | GEO | Homo sapiens | GSE73742 | Download (18,788) | |
| MEF2A | GM12878 | ENCODE | Homo sapiens | ENCSR000BKB | Download (34,610) | ||
| MEF2A | K-562 | ENCODE | Homo sapiens | ENCSR000BNV | Download (4,345) | ||
| MEF2A | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BVC | Download (11,715) | ||
| MEF2B | DOHH2 | GEO | Homo sapiens | GSE69558 | Download (7,240) | ||
| MEF2B | KARPAS422 | GEO | Homo sapiens | GSE69558 | Download (3,376) | ||
| MEF2B | OCI-Ly7 | GEO | Homo sapiens | GSE69558 | Download (660) | ||
| MEF2B | DLBCL | GEO | Homo sapiens | GSE110682 | Download (16,169) | ||
| MEF2B | tonsil | GEO | Homo sapiens | GSE110682 | Download (36,315) | ||
| MEF2B | GM12878 | ENCODE | Homo sapiens | ENCSR177VFS | Download (37,513) | ||
| MEF2C | GM12878 | ENCODE | Homo sapiens | ENCSR000BNG | Download (10,379) | ||
| MEF2C | HUVEC-C | GEO | Homo sapiens | GSE32644 | Download (277) | ||
| MEF2C | HUVEC-C | STATIN | GEO | Homo sapiens | GSE32644 | Download (242) | |
| MEF2C | MOLM-13 | GEO | Homo sapiens | GSE109492 | Download (100,747) | ||
| MEF2D | K-562 | ENCODE | Homo sapiens | ENCSR647ZXA | Download (8,187) | ||
| MEIS1 | HSPC | GEO | Homo sapiens | GSE26014 | Download (350) | ||
| MEIS1 | SEM | GEO | Homo sapiens | GSE38339 | Download (19,836) | ||
| MEIS1 | CHRF28811 | ENA | Homo sapiens | ERR063469 | Download (25,411) | ||
| MEIS2 | K-562 | ENCODE | Homo sapiens | ENCSR851BNE | Download (62,250) | ||
| MEN1 | SEM | GEO | Homo sapiens | GSE83671 | Download (2,184) | ||
| MEN1 | HT-1080 | GEO | Homo sapiens | GSE86504 | Download (241) | ||
| MEN1 | MCF-7 | GEO | Homo sapiens | GSE85317 | Download (18,843) | ||
| MEN1 | MCF-7 | E2 | GEO | Homo sapiens | GSE85317 | Download (8,476) | |
| MEN1 | ML-2 | GEO | Homo sapiens | GSE95511 | Download (1,408) | ||
| MGA | K-562 | ENCODE | Homo sapiens | ENCSR710WLO | Download (20,168) | ||
| MIER1 | K-562 | ENCODE | Homo sapiens | ENCSR426MDV | Download (17,702) | ||
| MIER2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR115PIK | Download (85,106) | ||
| MIER3 | Hep-G2 | ENCODE | Homo sapiens | ENCSR531VIJ | Download (80,300) | ||
| MITF | K-562 | ENCODE | Homo sapiens | ENCSR000FCB | Download (8,533) | ||
| MITF | K-562 | ENCODE | Homo sapiens | ENCSR797SWM | Download (35,291) | ||
| MITF | 501MEL | GEO | Homo sapiens | GSE61965 | Download (28,598) | ||
| MITF | 501MEL | GEO | Homo sapiens | GSE64137 | Download (36) | ||
| MITF | retina | pigment | GEO | Homo sapiens | GSE60024 | Download (2,772) | |
| MITF | melanocyte | BRAFV600E | GEO | Homo sapiens | GSE50681 | Download (24,500) | |
| MITF | melanocyte | CTR | GEO | Homo sapiens | GSE50681 | Download (31,912) | |
| MIXL1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR966PJY | Download (90,436) | ||
| MLL | MV4-11 | GEO | Homo sapiens | GSE79899 | Download (24,922) | ||
| MLL | THP-1 | GEO | Homo sapiens | GSE79899 | Download (22,644) | ||
| MLL4 | SW480 | GEO | Homo sapiens | GSE115985 | Download (41,047) | ||
| MLLT1 | MOLM-13 | GEO | Homo sapiens | GSE82116 | Download (7,319) | ||
| MLLT1 | MV4-11 | GEO | Homo sapiens | GSE82116 | Download (30,213) | ||
| MLLT1 | MV4-11 | GEO | Homo sapiens | GSE82116 | Download (8,948) | ||
| MLLT1 | K-562 | ENCODE | Homo sapiens | ENCSR675LRO | Download (18,205) | ||
| MLLT1 | MCF-7 | ENCODE | Homo sapiens | ENCSR427BBI | Download (5,454) | ||
| MLLT1 | GM12878 | ENCODE | Homo sapiens | ENCSR552XSN | Download (72,602) | ||
| MLLT1 | K-562 | ENCODE | Homo sapiens | ENCSR107GRP | Download (48,469) | ||
| MLLT3 | NPCS | GEO | Homo sapiens | GSE68332 | Download (110) | ||
| MLLT3 | THP-1 | GEO | Homo sapiens | GSE79899 | Download (20,357) | ||
| MLLT3 | CD34 | GEO | Homo sapiens | GSE54344 | Download (64) | ||
| MLX | Hep-G2 | ENCODE | Homo sapiens | ENCSR125DAD | Download (41,457) | ||
| MNT | Hep-G2 | ENCODE | Homo sapiens | ENCSR261EDU | Download (38,181) | ||
| MNT | Hep-G2 | ENCODE | Homo sapiens | ENCSR730TBC | Download (16,711) | ||
| MNT | K-562 | ENCODE | Homo sapiens | ENCSR390VGH | Download (28,004) | ||
| MNT | K-562 | ENCODE | Homo sapiens | ENCSR512NLO | Download (29,687) | ||
| MNT | K-562 | ENCODE | Homo sapiens | ENCSR979QYJ | Download (14,170) | ||
| MNT | MCF-7 | ENCODE | Homo sapiens | ENCSR663ZZZ | Download (34,347) | ||
| MORC2 | HeLa | V5-HUSH-KO | GEO | Homo sapiens | GSE95451 | Download (15,627) | |
| MORC2 | HeLa | V5-MORC2-KO | GEO | Homo sapiens | GSE95451 | Download (5,679) | |
| MORC2 | HeLa | V5-MORC2-KO-W505A-MORC2-mut | GEO | Homo sapiens | GSE95451 | Download (12,462) | |
| MORC2 | H9 | GEO | Homo sapiens | GSE95374 | Download (16,584) | ||
| MORC2 | K-562 | GEO | Homo sapiens | GSE95374 | Download (1,828) | ||
| MORC2 | K-562 | MPP8-KO | GEO | Homo sapiens | GSE95374 | Download (135) | |
| MORC2 | K-562 | TASOR-KO | GEO | Homo sapiens | GSE95374 | Download (176) | |
| MPHOSPH8 | H9 | GEO | Homo sapiens | GSE95374 | Download (17,063) | ||
| MRTFA | A-673-clone-Asp114 | GEO | Homo sapiens | GSE92738 | Download (10,866) | ||
| MRTFB | A-673-clone-Asp114 | GEO | Homo sapiens | GSE92738 | Download (71,935) | ||
| MRTFB | A-673-clone-Asp114 | dox | GEO | Homo sapiens | GSE94480 | Download (88,213) | |
| MTA1 | K-562 | ENCODE | Homo sapiens | ENCSR807BGP | Download (24,563) | ||
| MTA1 | MCF-7 | ENCODE | Homo sapiens | ENCSR348JOJ | Download (16,034) | ||
| MTA2 | MCF-7 | ENCODE | Homo sapiens | ENCSR551ZDZ | Download (1,056) | ||
| MTA2 | GM12878 | ENCODE | Homo sapiens | ENCSR293QAR | Download (58,782) | ||
| MTA2 | K-562 | ENCODE | Homo sapiens | ENCSR113LAS | Download (17,137) | ||
| MTA2 | K-562 | ENCODE | Homo sapiens | ENCSR411UYA | Download (37,554) | ||
| MTA3 | GM12878 | ENCODE | Homo sapiens | ENCSR000BRH | Download (26,173) | ||
| MTA3 | K-562 | ENCODE | Homo sapiens | ENCSR180NCY | Download (49,774) | ||
| MTA3 | K-562 | ENCODE | Homo sapiens | ENCSR914NEI | Download (42,543) | ||
| MTA3 | MCF-7 | ENCODE | Homo sapiens | ENCSR391KQC | Download (5,161) | ||
| MXD3 | Hep-G2 | ENCODE | Homo sapiens | ENCSR909GJR | Download (96,289) | ||
| MXD4 | Hep-G2 | ENCODE | Homo sapiens | ENCSR441KFW | Download (67,725) | ||
| MXI1 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZI | Download (5,875) | ||
| MXI1 | neural | ENCODE | Homo sapiens | ENCSR934NHU | Download (19,022) | ||
| MXI1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EDU | Download (18,671) | ||
| MXI1 | IMR-90 | ENCODE | Homo sapiens | ENCSR000EFE | Download (41,040) | ||
| MXI1 | K-562 | ENCODE | Homo sapiens | ENCSR000EGZ | Download (11,785) | ||
| MXI1 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000EIA | Download (45,201) | ||
| MXI1 | WA01 | ENCODE | Homo sapiens | ENCSR000EBR | Download (11,430) | ||
| MXI1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECU | Download (22,248) | ||
| MYB | CD4 | TH1 | GEO | Homo sapiens | GSE72266 | Download (37,257) | |
| MYB | CD4 | TH2 | GEO | Homo sapiens | GSE72266 | Download (11,474) | |
| MYB | GM12878 | ENCODE | Homo sapiens | ENCSR819ATC | Download (11,496) | ||
| MYB | U-937 | ATRA-treated_MYB | GEO | Homo sapiens | GSE98006 | Download (447) | |
| MYB | U-937 | vehicle-treated_MYB | GEO | Homo sapiens | GSE98006 | Download (1,488) | |
| MYB | Jurkat | GEO | Homo sapiens | GSE59657 | Download (63,691) | ||
| MYB | Loucy | GEO | Homo sapiens | GSE94000 | Download (36,385) | ||
| MYB | DU528 | GEO | Homo sapiens | GSE94000 | Download (31,386) | ||
| MYB | MOLT-3 | GEO | Homo sapiens | GSE59657 | Download (48,404) | ||
| MYB | MV4-11 | GEO | Homo sapiens | GSE94240 | Download (243) | ||
| MYB | MV4-11 | TG3 | GEO | Homo sapiens | GSE94240 | Download (256) | |
| MYB | PF-382 | GEO | Homo sapiens | GSE94000 | Download (26,208) | ||
| MYB | THP-1 | GEO | Homo sapiens | GSE90769 | Download (58,310) | ||
| MYB | THP-1 | OG86 | GEO | Homo sapiens | GSE90769 | Download (39,366) | |
| MYB101 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (635) |
| MYB107 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,230) |
| MYB108 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (128) |
| MYB118 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (3,655) |
| MYB27 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (242) |
| MYB3 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (17,731) |
| MYB3 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (12,939) |
| MYB33 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,329) |
| MYB3R-3 | Col-0 | Col-0_seedling | 9d | GEO | Arabidopsis thaliana | GSE60554 | Download (5,167) |
| MYB3R4 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,741) |
| MYB3R5 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (419) |
| MYB44 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (21,869) |
| MYB44 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (17,295) |
| MYB44 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,306) |
| MYB49 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (393) |
| MYB56 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,127) |
| MYB57 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (277) |
| MYB58 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,460) |
| MYB61 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (243) |
| MYB73 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,942) |
| MYB83 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (162) |
| MYB88 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (475) |
| MYB96 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (246) |
| MYB98 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,082) |
| MYB99 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (2,535) |
| MYBL2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BRO | Download (13,922) | ||
| MYBL2 | K-562 | ENCODE | Homo sapiens | ENCSR162IEM | Download (3,479) | ||
| MYBL2 | HMEC-1 | GEO | Homo sapiens | GSE62425 | Download (352) | ||
| MYC | hESC | GEO | Homo sapiens | GSE17917 | Download (4) | ||
| MYC | HFF | OHT | GEO | Homo sapiens | GSE65544 | Download (5,940) | |
| MYC | HFF | OHT_SHBPTF | GEO | Homo sapiens | GSE65544 | Download (5,292) | |
| MYC | IMEC | GEO | Homo sapiens | GSE70001 | Download (2,040) | ||
| MYC | IMEC | GEO | Homo sapiens | GSE86412 | Download (4,931) | ||
| MYC | IMEC | MYC | GEO | Homo sapiens | GSE86412 | Download (4,379) | |
| MYC | Jurkat | GEO | Homo sapiens | GSE83777 | Download (25,374) | ||
| MYC | LNCaP | MYC | GEO | Homo sapiens | GSE73994 | Download (32,252) | |
| MYC | LoVo | PHASEM | GEO | Homo sapiens | GSE51290 | Download (1,091) | |
| MYC | LoVo | PHASES | GEO | Homo sapiens | GSE51290 | Download (7,118) | |
| MYC | LS174T | BI8622 | GEO | Homo sapiens | GSE59223 | Download (4,248) | |
| MYC | LS174T | DMSO | GEO | Homo sapiens | GSE59223 | Download (924) | |
| MYC | PAVE | GEO | Homo sapiens | GSE47152 | Download (6,129) | ||
| MYC | Raji | GEO | Homo sapiens | GSE30726 | Download (11,591) | ||
| MYC | Ramos | GEO | Homo sapiens | GSE30726 | Download (6,326) | ||
| MYC | U2OS | GEO | Homo sapiens | GSE44672 | Download (5,454) | ||
| MYC | U2OS | Doxy | GEO | Homo sapiens | GSE44672 | Download (25,358) | |
| MYC | U2OS | Doxy | GEO | Homo sapiens | GSE77328 | Download (18,020) | |
| MYC | U2OS | EtOH | GEO | Homo sapiens | GSE77328 | Download (10,271) | |
| MYC | U2OS | HA-OmoMYCwt_Doxy | GEO | Homo sapiens | GSE77328 | Download (18,486) | |
| MYC | U2OS | HA-OmoMYCwt_EtOH | GEO | Homo sapiens | GSE77328 | Download (11,398) | |
| MYC | U2OS | GEO | Homo sapiens | GSE77356 | Download (5,706) | ||
| MYC | U2OS | Doxy | GEO | Homo sapiens | GSE77356 | Download (37,551) | |
| MYC | U2OS | SHMYC | GEO | Homo sapiens | GSE77356 | Download (6,679) | |
| MYC | U2OS | SHRCTR | GEO | Homo sapiens | GSE77356 | Download (30,852) | |
| MYC | DLD-1 | MYC-activated | GEO | Homo sapiens | GSE117240 | Download (12,374) | |
| MYC | HT-1080 | GEO | Homo sapiens | GSE86504 | Download (12,836) | ||
| MYC | HUVEC-C | GEO | Homo sapiens | GSE93030 | Download (305) | ||
| MYC | MDA-MB-231 | GEO | Homo sapiens | GSE48602 | Download (64) | ||
| MYC | MDA-MB-453 | DHT | GEO | Homo sapiens | GSE45201 | Download (19,486) | |
| MYC | NCI-H2171 | GEO | Homo sapiens | GSE36354 | Download (63,909) | ||
| MYC | NCI-H2171 | GEO | Homo sapiens | GSE41105 | Download (5,044) | ||
| MYC | P493-6 | GEO | Homo sapiens | GSE36354 | Download (3,936) | ||
| MYC | P493-6 | CMYC_1H | GEO | Homo sapiens | GSE36354 | Download (7,256) | |
| MYC | P493-6 | CMYC_24H | GEO | Homo sapiens | GSE36354 | Download (5,933) | |
| MYC | P493-6 | GEO | Homo sapiens | GSE42262 | Download (6,818) | ||
| MYC | P493-6 | SHCTR | GEO | Homo sapiens | GSE60223 | Download (18,358) | |
| MYC | P493-6 | SHTERC | GEO | Homo sapiens | GSE60223 | Download (16,887) | |
| MYC | P493-6 | SHTERT | GEO | Homo sapiens | GSE60223 | Download (1,913) | |
| MYC | MM1-S | DMSO | GEO | Homo sapiens | GSE36354 | Download (34,921) | |
| MYC | U-87MG | GEO | Homo sapiens | GSE36354 | Download (23,709) | ||
| MYC | MM1-S | JQ1 | GEO | Homo sapiens | GSE42161 | Download (3,390) | |
| MYC | GEN2-2 | GEO | Homo sapiens | GSE70275 | Download (10,210) | ||
| MYC | OSKM | GEO | Homo sapiens | GSE81899 | Download (248) | ||
| MYC | HCT-116 | GEO | Homo sapiens | GSE86556 | Download (6,324) | ||
| MYC | MDA-MB-231 | GEO | Homo sapiens | GSE95303 | Download (11,556) | ||
| MYC | iMEC | GEO | Homo sapiens | GSE86412 | Download (4,931) | ||
| MYC | MCF-10A | ENCODE | Homo sapiens | ENCSR000DOM | Download (20,215) | ||
| MYC | MCF-10A | ENCODE | Homo sapiens | ENCSR000DOS | Download (54,910) | ||
| MYC | MCF-7 | ENCODE | Homo sapiens | ENCSR000DMJ | Download (162,930) | ||
| MYC | MCF-7 | ENCODE | Homo sapiens | ENCSR000DMM | Download (102,371) | ||
| MYC | MCF-7 | ENCODE | Homo sapiens | ENCSR000DMP | Download (6,137) | ||
| MYC | MCF-7 | ENCODE | Homo sapiens | ENCSR000DMQ | Download (48,512) | ||
| MYC | P493-6 | Dpy30-shRNA | GEO | Homo sapiens | GSE101853 | Download (31,019) | |
| MYC | P493-6 | scramble-shRNA | GEO | Homo sapiens | GSE101853 | Download (36,807) | |
| MYC | WA01 | ENCODE | Homo sapiens | ENCSR000DLI | Download (997) | ||
| MYC | WA01 | ENCODE | Homo sapiens | ENCSR000EBY | Download (7,973) | ||
| MYC | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000DLN | Download (4,113) | ||
| MYC | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EZD | Download (42,524) | ||
| MYC | BE2C | SHCTR | GEO | Homo sapiens | GSE70098 | Download (15,592) | |
| MYC | BE2C | SHWDR5 | GEO | Homo sapiens | GSE70098 | Download (13,785) | |
| MYC | BJ | GEO | Homo sapiens | GSE36570 | Download (11,599) | ||
| MYC | BJ | INDUCED | GEO | Homo sapiens | GSE36570 | Download (31,997) | |
| MYC | BL41 | GEO | Homo sapiens | GSE30726 | Download (20,623) | ||
| MYC | BLUE1 | GEO | Homo sapiens | GSE30726 | Download (6,010) | ||
| MYC | breast | primary | GEO | Homo sapiens | GSE66252 | Download (2,041) | |
| MYC | CA46 | GEO | Homo sapiens | GSE30726 | Download (1,171) | ||
| MYC | CD34 | GEO | Homo sapiens | GSE85488 | Download (63,549) | ||
| MYC | CUTLL1 | GEO | Homo sapiens | GSE90716 | Download (5,517) | ||
| MYC | GP5D | GEO | Homo sapiens | GSE51234 | Download (22,935) | ||
| MYC | GP5D | SIRAD21 | GEO | Homo sapiens | GSE51234 | Download (8,804) | |
| MYC | H128 | GEO | Homo sapiens | GSE41105 | Download (40,310) | ||
| MYC | HeLa | GEO | Homo sapiens | GSE44672 | Download (11,356) | ||
| MYC | endothelial | umbilical-vein | ENCODE | Homo sapiens | ENCSR000DLU | Download (34,441) | |
| MYC | GM12878 | ENCODE | Homo sapiens | ENCSR000DKU | Download (8,035) | ||
| MYC | LCL | ENA | Homo sapiens | ERP004110 | Download (390,988) | ||
| MYC | NB4 | ENCODE | Homo sapiens | ENCSR000EHR | Download (7,750) | ||
| MYC | A-549 | ENCODE | Homo sapiens | ENCSR000DYC | Download (11,891) | ||
| MYC | Hep-G2 | ENCODE | Homo sapiens | ENCSR000DLR | Download (9,809) | ||
| MYC | K-562 | ENCODE | Homo sapiens | ENCSR000DLZ | Download (23,383) | ||
| MYC | K-562 | ENCODE | Homo sapiens | ENCSR000EGJ | Download (50,579) | ||
| MYC | K-562 | ENCODE | Homo sapiens | ENCSR000EGS | Download (25,810) | ||
| MYC | K-562 | ENCODE | Homo sapiens | ENCSR000EZU | Download (22,692) | ||
| MYC | K-562 | ENCODE | Homo sapiens | ENCSR000EZV | Download (11,762) | ||
| MYC | K-562 | ENCODE | Homo sapiens | ENCSR000FAG | Download (20,037) | ||
| MYC | K-562 | ENCODE | Homo sapiens | ENCSR000FAZ | Download (6,559) | ||
| MYCN | SHEP-21N | GEO | Homo sapiens | GSE80151 | Download (64,226) | ||
| MYCN | SHEP-21N | 24h | GEO | Homo sapiens | GSE80151 | Download (21,263) | |
| MYCN | SHEP-21N | DOX_0H | GEO | Homo sapiens | GSE80151 | Download (22,668) | |
| MYCN | SHEP-21N | DOX_24H | GEO | Homo sapiens | GSE80151 | Download (18,430) | |
| MYCN | SK-N-BE2-C | GEO | Homo sapiens | GSE80151 | Download (41,377) | ||
| MYCN | MYCN-3 | high | GEO | Homo sapiens | GSE83317 | Download (1,026) | |
| MYCN | MYCN-3 | low | GEO | Homo sapiens | GSE83317 | Download (20,175) | |
| MYCN | Kelly | GEO | Homo sapiens | GSE94782 | Download (41,832) | ||
| MYCN | Kelly | GEO | Homo sapiens | GSE94822 | Download (65,700) | ||
| MYCN | NGP | GEO | Homo sapiens | GSE94782 | Download (322,633) | ||
| MYCN | NB-1643 | GEO | Homo sapiens | GSE94782 | Download (17,160) | ||
| MYCN | SK-N-BE2-C | GEO | Homo sapiens | GSE94822 | Download (34,528) | ||
| MYCN | Kelly | GEO | Homo sapiens | GSE80151 | Download (8,013) | ||
| MYCN | NGP | GEO | Homo sapiens | GSE80151 | Download (17,875) | ||
| MYCN | RH4 | GEO | Homo sapiens | GSE83726 | Download (27,092) | ||
| MYCN | SH-EP | 2h | GEO | Homo sapiens | GSE80151 | Download (866) | |
| MYCN | SH-EP | 6h | GEO | Homo sapiens | GSE80151 | Download (1,862) | |
| MYCN | SH-SY5Y | ENA | Homo sapiens | ERP013415 | Download (399) | ||
| MYCN | SH-SY5Y | OVERMYCN_24H | ENA | Homo sapiens | ERP013415 | Download (132) | |
| MYCN | SH-SY5Y | OVERMYCN_48H | ENA | Homo sapiens | ERP013415 | Download (86) | |
| MYCN | SMS-KCN | ENA | Homo sapiens | ERP013415 | Download (145) | ||
| MYCN | SMS-KCNR | ENA | Homo sapiens | ERP013415 | Download (117) | ||
| MYCN | BE2C | GEO | Homo sapiens | GSE72640 | Download (658) | ||
| MYCN | BE2C | GEO | Homo sapiens | GSE80151 | Download (41,892) | ||
| MYF5 | Rh18 | GEO | Homo sapiens | GSE84628 | Download (19,152) | ||
| MYNN | HEK293 | ENCODE | Homo sapiens | ENCSR707BNG | Download (6,189) | ||
| MYNN | K-562 | ENCODE | Homo sapiens | ENCSR737LTZ | Download (41,028) | ||
| MYNN | HEK293 | GEO | Homo sapiens | GSE76494 | Download (3,528) | ||
| MYOD1 | myoblast | GEO | Homo sapiens | GSE50413 | Download (51,670) | ||
| MYOD1 | RHABDOMYOSARCOMA | GEO | Homo sapiens | GSE50413 | Download (64,155) | ||
| MYOD1 | RH30 | DMSO | GEO | Homo sapiens | GSE85169 | Download (50,475) | |
| MYOD1 | RH4 | GEO | Homo sapiens | GSE83726 | Download (46,221) | ||
| MYOG | RH30 | DMSO | GEO | Homo sapiens | GSE85169 | Download (20,213) | |
| MYOG | RH4 | GEO | Homo sapiens | GSE83726 | Download (23,996) | ||
| MYR2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,648) |
| MYR2 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (2,791) |
| MZF1 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (2,001) | ||
| MZF1 | HEK293 | ENCODE | Homo sapiens | ENCSR298QUH | Download (27,949) | ||
| NAB2 | HL-60 | PMA | GEO | Homo sapiens | GSE106359 | Download (2,297) | |
| NAC002 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (5,738) |
| NAC004 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,381) |
| NAC007 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,153) |
| NAC010 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,932) |
| NAC017 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,542) |
| NAC029 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,052) |
| NAC029 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (1,207) |
| NAC031 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,708) |
| NAC032 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (17,483) |
| NAC032 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (2,506) |
| NAC035 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,241) |
| NAC037 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,791) |
| NAC045 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,335) |
| NAC053 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,702) |
| NAC053 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (6,382) |
| NAC062 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,576) |
| NAC078 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (4,494) |
| NAC078 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (11,241) |
| NAC101 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (4,987) |
| NAC101 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (2,461) |
| NAC105 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (764) |
| NAC105 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (2,027) |
| NAC68 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (4,258) |
| NAC69 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,577) |
| NAC92 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,505) |
| NANOG | WA09 | GEO | Homo sapiens | GSE105028 | Download (45,201) | ||
| NANOG | WA09 | heat-shock | GEO | Homo sapiens | GSE105028 | Download (24,940) | |
| NANOG | GM23338 | ENCODE | Homo sapiens | ENCSR061DGF | Download (11,035) | ||
| NANOG | hESC | GEO | Homo sapiens | GSE18292 | Download (40,712) | ||
| NANOG | hESC | GEO | Homo sapiens | GSE20650 | Download (6,467) | ||
| NANOG | LNCaP | pNanog1_Dox | GEO | Homo sapiens | GSE74799 | Download (31,781) | |
| NANOG | LNCaP | pNanog8 | GEO | Homo sapiens | GSE74799 | Download (1,783) | |
| NANOG | LNCaP | pNanog8_Dox | GEO | Homo sapiens | GSE74799 | Download (31,321) | |
| NANOG | WA01 | ENCODE | Homo sapiens | ENCSR000BMT | Download (32,846) | ||
| NANOG | WA01 | ENA | Homo sapiens | ERP004238 | Download (40,186) | ||
| NANOG | WA01 | 3IL | ENA | Homo sapiens | ERP004238 | Download (67,531) | |
| NBN | MCF-7 | ENCODE | Homo sapiens | ENCSR591EBL | Download (302) | ||
| NBN | GM12878 | ENCODE | Homo sapiens | ENCSR278SQL | Download (54,070) | ||
| NBN | Hep-G2 | ENCODE | Homo sapiens | ENCSR210ZYL | Download (2,124) | ||
| NBN | K-562 | ENCODE | Homo sapiens | ENCSR085QEV | Download (37,333) | ||
| NCAPH2 | HEK293 | GEO | Homo sapiens | GSE97540 | Download (24,359) | ||
| NCOA1 | K-562 | ENCODE | Homo sapiens | ENCSR658WFQ | Download (23,159) | ||
| NCOA1 | K-562 | ENCODE | Homo sapiens | ENCSR711SNW | Download (939) | ||
| NCOA1 | K-562 | ENCODE | Homo sapiens | ENCSR931HNY | Download (12,888) | ||
| NCOA1 | MCF-7 | ENA | Homo sapiens | ERP000901 | Download (20,525) | ||
| NCOA1 | MCF-7 | E2 | ENA | Homo sapiens | ERP000901 | Download (11,859) | |
| NCOA1 | LS180 | GEO | Homo sapiens | GSE39277 | Download (4,772) | ||
| NCOA1 | LS180 | 125 | GEO | Homo sapiens | GSE39277 | Download (2,594) | |
| NCOA1 | U2OS | E2 | GEO | Homo sapiens | GSE26110 | Download (545) | |
| NCOA2 | THP-1 | Dex | GEO | Homo sapiens | GSE99887 | Download (317) | |
| NCOA2 | THP-1 | Dex | GEO | Homo sapiens | GSE99887 | Download (2,972) | |
| NCOA2 | K-562 | ENCODE | Homo sapiens | ENCSR349TZO | Download (1,212) | ||
| NCOA2 | K-562 | ENCODE | Homo sapiens | ENCSR803EKW | Download (1,132) | ||
| NCOA2 | MCF-7 | ENA | Homo sapiens | ERP000901 | Download (28,333) | ||
| NCOA2 | MCF-7 | E2 | ENA | Homo sapiens | ERP000901 | Download (48,631) | |
| NCOA3 | MCF-7 | ENCODE | Homo sapiens | ENCSR573OJP | Download (7,053) | ||
| NCOA3 | MCF-7 | ENCODE | Homo sapiens | ENCSR686SOV | Download (4,418) | ||
| NCOA3 | MCF-7 | ENA | Homo sapiens | ERP000901 | Download (154,558) | ||
| NCOA3 | MCF-7 | E2 | ENA | Homo sapiens | ERP000901 | Download (46,371) | |
| NCOA3 | H3396 | E2 | GEO | Homo sapiens | GSE32349 | Download (2,795) | |
| NCOA4 | erythroblast | GC-1 | GEO | Homo sapiens | GSE102584 | Download (213) | |
| NCOA4 | K-562 | ENCODE | Homo sapiens | ENCSR119ULQ | Download (2,074) | ||
| NCOA6 | K-562 | ENCODE | Homo sapiens | ENCSR168CEE | Download (1,600) | ||
| NCOR1 | K-562 | ENCODE | Homo sapiens | ENCSR000ATY | Download (819) | ||
| NCOR1 | K-562 | ENCODE | Homo sapiens | ENCSR298JCG | Download (5,788) | ||
| NCOR1 | K-562 | ENCODE | Homo sapiens | ENCSR798ILC | Download (33,592) | ||
| NCOR1 | K-562 | ENCODE | Homo sapiens | ENCSR910JAI | Download (17,729) | ||
| NCOR1 | HEK293T | SICTR | GEO | Homo sapiens | GSE35197 | Download (13,124) | |
| NCOR1 | HEK293T | SIGSP2 | GEO | Homo sapiens | GSE35197 | Download (1,754) | |
| NCOR1 | LS180 | GEO | Homo sapiens | GSE39277 | Download (37,930) | ||
| NCOR1 | LS180 | 125 | GEO | Homo sapiens | GSE39277 | Download (39,101) | |
| NCOR1 | OCI-Ly1 | GEO | Homo sapiens | GSE29282 | Download (8,511) | ||
| NCOR2 | LS180 | GEO | Homo sapiens | GSE39277 | Download (17,156) | ||
| NCOR2 | LS180 | 125 | GEO | Homo sapiens | GSE39277 | Download (29,423) | |
| NCOR2 | OCI-Ly1 | GEO | Homo sapiens | GSE29282 | Download (7,715) | ||
| NCOR2 | B-cell | GERMINAL_CENTER | GEO | Homo sapiens | GSE43350 | Download (10,519) | |
| NELFA | HeLa | 1h-Flavo-0-H2O2 | GEO | Homo sapiens | GSE93931 | Download (9,783) | |
| NELFA | HeLa | 1h_Flavo-35min-H2O2 | GEO | Homo sapiens | GSE93931 | Download (11,907) | |
| NELFA | HeLa | Flavo-0-H2O2 | GEO | Homo sapiens | GSE100742 | Download (9,950) | |
| NELFA | HeLa | Flavo-10min-H2O2 | GEO | Homo sapiens | GSE100742 | Download (4,228) | |
| NELFA | HeLa | Flavo-35min-H2O2 | GEO | Homo sapiens | GSE100742 | Download (12,149) | |
| NELFA | HeLa | Flavo-PJ34-0-H2O2 | GEO | Homo sapiens | GSE100742 | Download (6,027) | |
| NELFA | HeLa | Flavo-PJ34-10min-H2O2 | GEO | Homo sapiens | GSE100742 | Download (7,052) | |
| NELFA | BT-474 | ENA | Homo sapiens | ERP010664 | Download (5,560) | ||
| NELFA | BT-474 | INHHDAC | ENA | Homo sapiens | ERP010664 | Download (12,056) | |
| NELFA | HeLa | 40min-Flavo-0-H2O2 | GEO | Homo sapiens | GSE93931 | Download (5,162) | |
| NELFA | HeLa | 40min-Flavo-10min-H2O2 | GEO | Homo sapiens | GSE93931 | Download (4,228) | |
| NELFA | HeLa | 40min-Flavo-PJ34-0-H2O2 | GEO | Homo sapiens | GSE93931 | Download (6,027) | |
| NELFA | HeLa | 40min-Flavo-PJ34-10min-H2O2 | GEO | Homo sapiens | GSE93931 | Download (7,052) | |
| NEUROD1 | NCI-H524 | GEO | Homo sapiens | GSE69394 | Download (3,672) | ||
| NEUROD1 | K-562 | ENCODE | Homo sapiens | ENCSR986CDX | Download (48,142) | ||
| NEUROD1 | MCF-7 | ENCODE | Homo sapiens | ENCSR006WUS | Download (7,499) | ||
| NEUROD1 | D283-Med | GEO | Homo sapiens | GSE92582 | Download (74,307) | ||
| NEUROD1 | D283-Med | shGFP | GEO | Homo sapiens | GSE92582 | Download (103,679) | |
| NEUROD1 | D283-Med | shNEUROD1-1154 | GEO | Homo sapiens | GSE92582 | Download (91,308) | |
| NEUROD1 | D283-Med | shNEUROD1-1579 | GEO | Homo sapiens | GSE92582 | Download (55,199) | |
| NEUROD1 | D341-Med | GEO | Homo sapiens | GSE92582 | Download (82,645) | ||
| NEUROD1 | D341-Med | shGFP | GEO | Homo sapiens | GSE92582 | Download (65,688) | |
| NEUROD1 | D341-Med | shNEUROD1-1154 | GEO | Homo sapiens | GSE92582 | Download (29,597) | |
| NEUROD1 | D341-Med | shNEUROD1-1579 | GEO | Homo sapiens | GSE92582 | Download (40,155) | |
| NEUROG2 | MRC-5 | N_02DPT | GEO | Homo sapiens | GSE75910 | Download (25,415) | |
| NEUROG2 | MRC-5 | N_05DPT | GEO | Homo sapiens | GSE75910 | Download (20,203) | |
| NEUROG2 | MRC-5 | N_1DPT | GEO | Homo sapiens | GSE75910 | Download (28,311) | |
| NEUROG2 | MRC-5 | NFD_02DPT | GEO | Homo sapiens | GSE75910 | Download (39,590) | |
| NEUROG2 | MRC-5 | NFD_05DPT | GEO | Homo sapiens | GSE75910 | Download (40,323) | |
| NEUROG2 | MRC-5 | NFD_1DPT | GEO | Homo sapiens | GSE75910 | Download (43,946) | |
| NFATC1 | HUVEC-C | GEO | Homo sapiens | GSE49428 | Download (14,520) | ||
| NFATC1 | HUVEC-C | VEGF | GEO | Homo sapiens | GSE49428 | Download (16,120) | |
| NFATC1 | GM12878 | ENCODE | Homo sapiens | ENCSR000BQL | Download (34,739) | ||
| NFATC3 | GM12878 | ENCODE | Homo sapiens | ENCSR437GBJ | Download (51,225) | ||
| NFATC3 | K-562 | ENCODE | Homo sapiens | ENCSR051OUX | Download (16,025) | ||
| NFATC3 | Hep-G2 | GEO | Homo sapiens | GSE97661 | Download (27,214) | ||
| NFATC3 | K-562 | ENCODE | Homo sapiens | ENCSR670FDA | Download (6,591) | ||
| NFE2 | erythroid | R3R4 | GEO | Homo sapiens | GSE43625 | Download (28,478) | |
| NFE2 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZY | Download (190) | ||
| NFE2 | K-562 | ENCODE | Homo sapiens | ENCSR000FAF | Download (5,093) | ||
| NFE2 | K-562 | ENCODE | Homo sapiens | ENCSR000FCC | Download (47,245) | ||
| NFE2 | K-562 | ENCODE | Homo sapiens | ENCSR552YGL | Download (31,920) | ||
| NFE2 | ProEs | GEO | Homo sapiens | GSE59087 | Download (71,430) | ||
| NFE2L1 | K-562 | ENCODE | Homo sapiens | ENCSR632SHZ | Download (4,328) | ||
| NFE2L2 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR707IUN | Download (28,149) | ||
| NFE2L2 | BEAS-2B | GEO | Homo sapiens | GSE75812 | Download (204) | ||
| NFE2L2 | A-549 | GEO | Homo sapiens | GSE113497 | Download (21,751) | ||
| NFE2L2 | A-375 | A771726 | GEO | Homo sapiens | GSE57431 | Download (5,654) | |
| NFE2L2 | A-375 | DMSO | GEO | Homo sapiens | GSE57431 | Download (11,031) | |
| NFE2L2 | HAEC | oxpapc_4h | GEO | Homo sapiens | GSE89970 | Download (2,263) | |
| NFE2L2 | A-549 | ENCODE | Homo sapiens | ENCSR584GHV | Download (10,665) | ||
| NFE2L2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR488EES | Download (18,033) | ||
| NFE2L2 | IMR-90 | ENCODE | Homo sapiens | ENCSR197WGI | Download (32,792) | ||
| NFE2L2 | K-562 | ENCODE | Homo sapiens | ENCSR485GKE | Download (55,982) | ||
| NFIA | Hep-G2 | ENCODE | Homo sapiens | ENCSR226QQM | Download (98,083) | ||
| NFIA | Hep-G2 | GEO | Homo sapiens | GSE97661 | Download (8,739) | ||
| NFIA | K-562 | GEO | Homo sapiens | GSE97661 | Download (24,254) | ||
| NFIB | MCF-7 | ENCODE | Homo sapiens | ENCSR582ZOA | Download (19,496) | ||
| NFIB | MCF-7 | ENCODE | Homo sapiens | ENCSR702BYX | Download (31,986) | ||
| NFIC | K-562 | ENCODE | Homo sapiens | ENCSR796ITY | Download (53,215) | ||
| NFIC | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BSV | Download (59,240) | ||
| NFIC | GM12878 | ENCODE | Homo sapiens | ENCSR000BRN | Download (43,357) | ||
| NFIC | Ishikawa | ENCODE | Homo sapiens | ENCSR000BUT | Download (47,821) | ||
| NFIC | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BQX | Download (14,666) | ||
| NFIC | Hep-G2 | GEO | Homo sapiens | GSE108514 | Download (11,870) | ||
| NFIL3 | Hep-G2 | ENCODE | Homo sapiens | ENCSR201GGK | Download (108,937) | ||
| NFIL3 | Hep-G2 | GEO | Homo sapiens | GSE97661 | Download (6,029) | ||
| NFKB1 | L1236 | GEO | Homo sapiens | GSE63736 | Download (56,732) | ||
| NFKB1 | MCF10A-Er-Src | EtOH | GEO | Homo sapiens | GSE115597 | Download (9,772) | |
| NFKB1 | MCF10A-Er-Src | TAM | GEO | Homo sapiens | GSE115597 | Download (54,747) | |
| NFKB2 | L1236 | GEO | Homo sapiens | GSE63736 | Download (25,374) | ||
| NFKBIA | dermal | GEO | Homo sapiens | GSE30082 | Download (1,787) | ||
| NFKBIZ | Hep-G2 | ENCODE | Homo sapiens | ENCSR235OVI | Download (63,698) | ||
| NFRKB | HEK293T | ENCODE | Homo sapiens | ENCSR997GPO | Download (642) | ||
| NFRKB | Hep-G2 | ENCODE | Homo sapiens | ENCSR145BHD | Download (2,090) | ||
| NFRKB | K-562 | ENCODE | Homo sapiens | ENCSR657EOF | Download (20,502) | ||
| NFRKB | K-562 | ENCODE | Homo sapiens | ENCSR996ESX | Download (3,151) | ||
| NFXL1 | MCF-7 | ENCODE | Homo sapiens | ENCSR417DKD | Download (458) | ||
| NFXL1 | GM12878 | ENCODE | Homo sapiens | ENCSR746XEG | Download (7,096) | ||
| NFXL1 | K-562 | ENCODE | Homo sapiens | ENCSR085DDI | Download (5,545) | ||
| NFYA | GM12878 | ENCODE | Homo sapiens | ENCSR000DNN | Download (1,023) | ||
| NFYA | K-562 | ENCODE | Homo sapiens | ENCSR000EGR | Download (4,798) | ||
| NFYA | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000DNS | Download (7,772) | ||
| NFYA | K-562 | GEO | Homo sapiens | GSE26439 | Download (26,336) | ||
| NFYB | K-562 | GEO | Homo sapiens | GSE26439 | Download (15,253) | ||
| NFYB | GM12878 | ENCODE | Homo sapiens | ENCSR000DNM | Download (8,893) | ||
| NFYB | K-562 | ENCODE | Homo sapiens | ENCSR000EGQ | Download (13,231) | ||
| NFYB | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000DNR | Download (8,235) | ||
| NFYB2 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (28,697) |
| NFYB2 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (26,676) |
| NFYB9 | Ws-0 | Ws-0_seeds-bent-cotyledon-stage | GEO | Arabidopsis thaliana | GSE99587 | Download (18,861) | |
| NFYC | Hep-G2 | ENCODE | Homo sapiens | ENCSR569ARC | Download (41,020) | ||
| NFYC2 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (19,865) |
| NFYC2 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (15,345) |
| NGA4 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (475) |
| NIPBL | HCT-116 | RAD21-mAC | GEO | Homo sapiens | GSE104888 | Download (29,774) | |
| NIPBL | HCT-116 | RAD21-mAC_500uM_auxin | GEO | Homo sapiens | GSE104888 | Download (33,525) | |
| NIPBL | GM12878 | GEO | Homo sapiens | GSE93080 | Download (25,070) | ||
| NIPBL | GP5D | GEO | Homo sapiens | GSE51234 | Download (26,531) | ||
| NIPBL | GP5D | SIRAD21 | GEO | Homo sapiens | GSE51234 | Download (4,500) | |
| NIPBL | hESC | GEO | Homo sapiens | GSE64758 | Download (3,857) | ||
| NIPBL | hESC | activin | GEO | Homo sapiens | GSE64758 | Download (4,367) | |
| NIPBL | hESC | WNT3A | GEO | Homo sapiens | GSE64758 | Download (4,106) | |
| NIPBL | hESC | WNT3A_ACTIVIN | GEO | Homo sapiens | GSE64758 | Download (7,923) | |
| NIPBL | LCL | GEO | Homo sapiens | GSE38395 | Download (11,191) | ||
| NIPBL | A-549 | GEO | Homo sapiens | GSE76893 | Download (13,317) | ||
| NIPBL | Hep-G2 | GEO | Homo sapiens | GSE76893 | Download (7,549) | ||
| NIPBL | MCF-7 | GEO | Homo sapiens | GSE76893 | Download (6,859) | ||
| NIPBL | WA09 | GEO | Homo sapiens | GSE105028 | Download (4,138) | ||
| NIPBL | WA09 | heat-shock | GEO | Homo sapiens | GSE105028 | Download (2,071) | |
| NKX2-1 | H9 | derived-cIN | GEO | Homo sapiens | GSE99937 | Download (53,419) | |
| NKX2-1 | A-549 | GEO | Homo sapiens | GSE86957 | Download (131,266) | ||
| NKX2-1 | NCI-H1819 | GEO | Homo sapiens | GSE39998 | Download (11,238) | ||
| NKX2-1 | NCI-H2087 | GEO | Homo sapiens | GSE39998 | Download (13,635) | ||
| NKX2-1 | NCI-H3122 | GEO | Homo sapiens | GSE39998 | Download (61,521) | ||
| NKX2-2 | islet | ENA | Homo sapiens | ERP004003 | Download (137,600) | ||
| NKX2-5 | hESC | ab3584 | GEO | Homo sapiens | GSE89457 | Download (18,940) | |
| NKX2-5 | hESC | sc-14033 | GEO | Homo sapiens | GSE89457 | Download (11,097) | |
| NKX3-1 | LNCaP | DHT | GEO | Homo sapiens | GSE28264 | Download (10,241) | |
| NKX3-1 | LNCaP | ETOH | GEO | Homo sapiens | GSE28264 | Download (3,186) | |
| NKX3-1 | LNCaP | GEO | Homo sapiens | GSE40269 | Download (93,666) | ||
| NKX3-1 | islet | ENA | Homo sapiens | ERP004003 | Download (71,863) | ||
| NME2 | A-549 | IND | GEO | Homo sapiens | GSE40300 | Download (129,753) | |
| NMYC | IMR-5 | CD532 | GEO | Homo sapiens | GSE78957 | Download (6,846) | |
| NMYC | IMR-5 | DMSO | GEO | Homo sapiens | GSE78957 | Download (6,568) | |
| NONO | Hep-G2 | ENCODE | Homo sapiens | ENCSR476BQA | Download (3,608) | ||
| NONO | Hep-G2 | ENCODE | Homo sapiens | ENCSR923UTX | Download (20,252) | ||
| NONO | K-562 | ENCODE | Homo sapiens | ENCSR010KFT | Download (14,318) | ||
| NONO | K-562 | ENCODE | Homo sapiens | ENCSR415TXN | Download (7,595) | ||
| NONO | K-562 | ENCODE | Homo sapiens | ENCSR886RYH | Download (6,034) | ||
| NONO | MCF-7 | ENCODE | Homo sapiens | ENCSR912NMR | Download (4,522) | ||
| NOTCH1 | CD34 | GEO | Homo sapiens | GSE63010 | Download (5,632) | ||
| NOTCH1 | CUTLL1 | GEO | Homo sapiens | GSE29600 | Download (19,969) | ||
| NOTCH1 | REC-1 | GSI-mock-washout | GEO | Homo sapiens | GSE97541 | Download (118) | |
| NOTCH1 | REC-1 | GSI-washout | GEO | Homo sapiens | GSE97541 | Download (2,242) | |
| NOTCH1 | REC-1 | GEO | Homo sapiens | GSE97541 | Download (1,305) | ||
| NOTCH1 | SP-49 | GEO | Homo sapiens | GSE97541 | Download (282) | ||
| NOTCH1 | HPBALL | GEO | Homo sapiens | GSE39263 | Download (59,881) | ||
| NOTCH1 | GSC8-11 | 12d-das | GEO | Homo sapiens | GSE74557 | Download (453) | |
| NOTCH1 | GSC8-11 | dasatinib | GEO | Homo sapiens | GSE74557 | Download (2,547) | |
| NOTCH3 | TALL-1 | DMSO | GEO | Homo sapiens | GSE104261 | Download (6,733) | |
| NOTCH3 | TALL-1 | GSI | GEO | Homo sapiens | GSE104261 | Download (17,362) | |
| NR0B1 | NCI-H460 | GEO | Homo sapiens | GSE89569 | Download (987) | ||
| NR1H2 | HT29 | GW3965_2H | GEO | Homo sapiens | GSE77039 | Download (15,625) | |
| NR1H2 | HT29 | GW3965_48H | GEO | Homo sapiens | GSE77039 | Download (38,002) | |
| NR1H3 | SGBS | GEO | Homo sapiens | GSE41629 | Download (1,254) | ||
| NR1H3 | THP-1 | GEO | Homo sapiens | GSE28319 | Download (331) | ||
| NR1H3 | THP-1 | T09 | GEO | Homo sapiens | GSE28319 | Download (317) | |
| NR2C1 | K-562 | ENCODE | Homo sapiens | ENCSR742IDN | Download (11,519) | ||
| NR2C1 | GM12878 | ENCODE | Homo sapiens | ENCSR784VIQ | Download (7,284) | ||
| NR2C1 | K-562 | ENCODE | Homo sapiens | ENCSR178DEG | Download (18,209) | ||
| NR2C2 | GM12878 | ENCODE | Homo sapiens | ENCSR000EUL | Download (362) | ||
| NR2C2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EVS | Download (3,014) | ||
| NR2C2 | K-562 | ENCODE | Homo sapiens | ENCSR750LYM | Download (16,778) | ||
| NR2C2 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EVN | Download (1,800) | ||
| NR2C2 | erythroid | D8 | GEO | Homo sapiens | GSE54759 | Download (323) | |
| NR2C2 | HeLa | GEO | Homo sapiens | GSE46237 | Download (912) | ||
| NR2C2 | WI-38VA13 | GEO | Homo sapiens | GSE46237 | Download (1,726) | ||
| NR2F1 | GM12878 | ENCODE | Homo sapiens | ENCSR514VYD | Download (41,844) | ||
| NR2F1 | MCF-7 | GEO | Homo sapiens | GSE41561 | Download (113,958) | ||
| NR2F1 | K-562 | ENCODE | Homo sapiens | ENCSR970NKQ | Download (49,039) | ||
| NR2F2 | MCF-7 | ENCODE | Homo sapiens | ENCSR000BUY | Download (59,367) | ||
| NR2F2 | HeLa | GEO | Homo sapiens | GSE46237 | Download (4,942) | ||
| NR2F2 | liver | ENCODE | Homo sapiens | ENCSR168SMX | Download (63,570) | ||
| NR2F2 | liver | ENCODE | Homo sapiens | ENCSR338MMB | Download (43,459) | ||
| NR2F2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BVM | Download (22,782) | ||
| NR2F2 | K-562 | ENCODE | Homo sapiens | ENCSR000BRS | Download (63,669) | ||
| NR2F2 | WI-38VA13 | GEO | Homo sapiens | GSE46237 | Download (5,093) | ||
| NR2F2 | uterus | STROMA_ENDOMETRIUM | GEO | Homo sapiens | GSE52008 | Download (26,927) | |
| NR2F6 | Hep-G2 | ENCODE | Homo sapiens | ENCSR518WPL | Download (41,794) | ||
| NR2F6 | Hep-G2 | ENCODE | Homo sapiens | ENCSR798IJO | Download (97,692) | ||
| NR2F6 | K-562 | ENCODE | Homo sapiens | ENCSR707QWA | Download (33,451) | ||
| NR3C1 | K-562 | GLUCC | ENA | Homo sapiens | ERP007081 | Download (18,649) | |
| NR3C1 | MCF-10A | DEX_20min | GEO | Homo sapiens | GSE102355 | Download (5,897) | |
| NR3C1 | MCF-10A | DEX_60min | GEO | Homo sapiens | GSE102355 | Download (12,221) | |
| NR3C1 | MCF-10A | EGF_DEX_20min | GEO | Homo sapiens | GSE102355 | Download (11,806) | |
| NR3C1 | MCF-10A | EGF_DEX_60min | GEO | Homo sapiens | GSE102355 | Download (9,215) | |
| NR3C1 | MCF-7 | GEO | Homo sapiens | GSE104399 | Download (7,842) | ||
| NR3C1 | breast | tumor_Male_1 | GEO | Homo sapiens | GSE104399 | Download (5,982) | |
| NR3C1 | breast | tumor_Male_12 | GEO | Homo sapiens | GSE104399 | Download (1,912) | |
| NR3C1 | breast | tumor_Male_15 | GEO | Homo sapiens | GSE104399 | Download (10,996) | |
| NR3C1 | breast | tumor_Male_21 | GEO | Homo sapiens | GSE104399 | Download (24,140) | |
| NR3C1 | breast | tumor_Male_9 | GEO | Homo sapiens | GSE104399 | Download (5,395) | |
| NR3C1 | GM12878 | ENCODE | Homo sapiens | ENCSR904YPP | Download (15,752) | ||
| NR3C1 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BJC | Download (56,562) | ||
| NR3C1 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BLB | Download (8,158) | ||
| NR3C1 | U2OS | GLUCC | ENA | Homo sapiens | ERP007081 | Download (50,776) | |
| NR3C1 | A-549 | ENCODE | Homo sapiens | ENCSR000BHE | Download (15,614) | ||
| NR3C1 | A-549 | ENCODE | Homo sapiens | ENCSR000BHF | Download (79,632) | ||
| NR3C1 | A-549 | ENCODE | Homo sapiens | ENCSR000BHG | Download (37,434) | ||
| NR3C1 | A-549 | ENCODE | Homo sapiens | ENCSR000BIO | Download (240) | ||
| NR3C1 | A-549 | ENCODE | Homo sapiens | ENCSR000BJR | Download (77,012) | ||
| NR3C1 | A-549 | ENCODE | Homo sapiens | ENCSR000BJT | Download (8,579) | ||
| NR3C1 | A-549 | ENCODE | Homo sapiens | ENCSR116TFA | Download (12,053) | ||
| NR3C1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEV | Download (351) | ||
| NR3C1 | IMR-90 | GLUCC | ENA | Homo sapiens | ERP007081 | Download (42,052) | |
| NR3C1 | IMR-90 | ENA | Homo sapiens | ERP007093 | Download (44,780) | ||
| NR3C1 | K-562 | ENCODE | Homo sapiens | ENCSR325DER | Download (654) | ||
| NR3C1 | K-562 | ENCODE | Homo sapiens | ENCSR494UQJ | Download (1,636) | ||
| NR3C1 | HeLa-B2 | DMSO | GEO | Homo sapiens | GSE24518 | Download (451) | |
| NR3C1 | HeLa-B2 | GRKD_DMSO | GEO | Homo sapiens | GSE24518 | Download (14,499) | |
| NR3C1 | HeLa-B2 | GRKD_TA_TNFA | GEO | Homo sapiens | GSE24518 | Download (15,536) | |
| NR3C1 | HeLa-B2 | P65KD_DMSO | GEO | Homo sapiens | GSE24518 | Download (9,108) | |
| NR3C1 | HeLa-B2 | P65KD_TA_TNFA | GEO | Homo sapiens | GSE24518 | Download (29,090) | |
| NR3C1 | HeLa-B2 | TA | GEO | Homo sapiens | GSE24518 | Download (11,131) | |
| NR3C1 | HeLa-B2 | TA_TNFA | GEO | Homo sapiens | GSE24518 | Download (9,264) | |
| NR3C1 | HeLa-B2 | TNFA | GEO | Homo sapiens | GSE24518 | Download (438) | |
| NR3C1 | MM1-S | GEO | Homo sapiens | GSE63209 | Download (28) | ||
| NR3C1 | MM1-S | DEX | GEO | Homo sapiens | GSE63209 | Download (21) | |
| NR3C1 | RS4-11 | DEX | GEO | Homo sapiens | GSE71616 | Download (1,123) | |
| NR3C1 | THP-1 | GEO | Homo sapiens | GSE99887 | Download (871) | ||
| NR3C1 | THP-1 | Dex | GEO | Homo sapiens | GSE99887 | Download (16,037) | |
| NR3C1 | HASM2 | Dex | GEO | Homo sapiens | GSE95632 | Download (99,252) | |
| NR3C1 | HASM2 | Dex-GFP | GEO | Homo sapiens | GSE95632 | Download (86,177) | |
| NR3C1 | HASM2 | Dex-KLF115 | GEO | Homo sapiens | GSE95632 | Download (95,115) | |
| NR3C1 | HASM2 | EtOH | GEO | Homo sapiens | GSE95632 | Download (90,574) | |
| NR3C1 | hMSC | DMI | GEO | Homo sapiens | GSE68864 | Download (5,682) | |
| NR3C1 | Ishikawa | GEO | Homo sapiens | GSE109891 | Download (1,768) | ||
| NR3C1 | Ishikawa | Dex | GEO | Homo sapiens | GSE109891 | Download (12,076) | |
| NR3C1 | Ishikawa | Dex_E2 | GEO | Homo sapiens | GSE109891 | Download (8,085) | |
| NR3C1 | LCL | DEX | GEO | Homo sapiens | GSE45638 | Download (3,173) | |
| NR3C1 | LCL | ETOH | GEO | Homo sapiens | GSE45638 | Download (158) | |
| NR3C1 | LNCaP | 1F5 | GEO | Homo sapiens | GSE30623 | Download (10,779) | |
| NR3C1 | LNCaP | 1F5_SIFOXA1 | GEO | Homo sapiens | GSE30623 | Download (16,661) | |
| NR3C1 | U2OS | siBRMsiHic5 | GEO | Homo sapiens | GSE109383 | Download (8,369) | |
| NR3C1 | U2OS | siHic5siNS | GEO | Homo sapiens | GSE109383 | Download (17,034) | |
| NR3C1 | U2OS | SHHIC5 | GEO | Homo sapiens | GSE65847 | Download (55,014) | |
| NR3C1 | U2OS | SHNS | GEO | Homo sapiens | GSE65847 | Download (16,961) | |
| NR3C1 | ZR751 | GEO | Homo sapiens | GSE72249 | Download (4,607) | ||
| NR3C1 | ZR751 | DEX | GEO | Homo sapiens | GSE72249 | Download (19,479) | |
| NR3C1 | MCF-7 | GEO | Homo sapiens | GSE72249 | Download (791) | ||
| NR3C1 | MCF-7 | DEX | GEO | Homo sapiens | GSE72249 | Download (13,790) | |
| NR3C1 | MCF-7 | E2_Dex | GEO | Homo sapiens | GSE81510 | Download (41,127) | |
| NR3C1 | MCF-7 | ICI_Dex | GEO | Homo sapiens | GSE81510 | Download (34,297) | |
| NR3C1 | MCF-7 | E2_Dex | GEO | Homo sapiens | GSE81510 | Download (18,754) | |
| NR3C1 | MCF-7 | ICI_Dex | GEO | Homo sapiens | GSE81510 | Download (24,531) | |
| NR3C1 | NALM-6 | GEO | Homo sapiens | GSE67046 | Download (15,825) | ||
| NR3C1 | NALM-6 | CASP1 | GEO | Homo sapiens | GSE67046 | Download (5,337) | |
| NR3C1 | T-47D | GEO | Homo sapiens | GSE72249 | Download (159) | ||
| NR3C1 | T-47D | DEX | GEO | Homo sapiens | GSE72249 | Download (733) | |
| NR4A1 | Kasumi-1 | GEO | Homo sapiens | GSE79491 | Download (33,666) | ||
| NR4A1 | K-562 | ENCODE | Homo sapiens | ENCSR000DJW | Download (1,508) | ||
| NR4A1 | K-562 | ENCODE | Homo sapiens | ENCSR130PDE | Download (13,502) | ||
| NR5A2 | MCF-7 | GEO | Homo sapiens | GSE47027 | Download (4,571) | ||
| NRF1 | Namalwa | GEO | Homo sapiens | GSE53133 | Download (6,890) | ||
| NRF1 | HMEC-1 | GEO | Homo sapiens | GSE67867 | Download (30,352) | ||
| NRF1 | T-47D | HAEGIN2 | GEO | Homo sapiens | GSE59935 | Download (13,470) | |
| NRF1 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZO | Download (3,564) | ||
| NRF1 | HeLa | dC9Sun-D3A_TMEM206 | GEO | Homo sapiens | GSE107607 | Download (7,556) | |
| NRF1 | HeLa | dC9Sun-mCherry_TMEM206 | GEO | Homo sapiens | GSE107607 | Download (4,804) | |
| NRF1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEH | Download (2,217) | ||
| NRF1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR853ADA | Download (17,060) | ||
| NRF1 | K-562 | ENCODE | Homo sapiens | ENCSR000EHH | Download (5,110) | ||
| NRF1 | K-562 | ENCODE | Homo sapiens | ENCSR494TDU | Download (18,184) | ||
| NRF1 | K-562 | ENCODE | Homo sapiens | ENCSR837EYC | Download (25,510) | ||
| NRF1 | K-562 | ENCODE | Homo sapiens | ENCSR998AJK | Download (19,962) | ||
| NRF1 | MCF-7 | ENCODE | Homo sapiens | ENCSR135ANT | Download (5,710) | ||
| NRF1 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000EHZ | Download (9,505) | ||
| NRF1 | WA01 | ENCODE | Homo sapiens | ENCSR000ECC | Download (4,473) | ||
| NRF1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EDJ | Download (3,174) | ||
| NRF1 | HeLa-S3 | GEO | Homo sapiens | GSE108856 | Download (36,805) | ||
| NRF1 | HCC1954 | GEO | Homo sapiens | GSE67867 | Download (27,898) | ||
| NRF1 | Hep-G2 | GEO | Homo sapiens | GSE97661 | Download (9,031) | ||
| NRF1 | K-562 | Ab_R157-1-3D4 | GEO | Homo sapiens | GSE97661 | Download (19,793) | |
| NRF1 | K-562 | Ab_R157-1-3H1 | GEO | Homo sapiens | GSE97661 | Download (8,992) | |
| NRF1 | K-562 | Ab_R157-1-3H3 | GEO | Homo sapiens | GSE97661 | Download (16,388) | |
| NRF1 | MCF-7 | Ab_R157-1-3D4 | GEO | Homo sapiens | GSE97661 | Download (10,077) | |
| NRF1 | MCF-7 | Ab_R157-1-3H1 | GEO | Homo sapiens | GSE97661 | Download (4,502) | |
| NRIP1 | MCF-7 | ENA | Homo sapiens | ERP005838 | Download (64,316) | ||
| NRIP1 | MCF-7 | E2 | ENA | Homo sapiens | ERP005838 | Download (78,403) | |
| NSD2 | K-562 | ENCODE | Homo sapiens | ENCSR000AVE | Download (7,348) | ||
| NTL8 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,832) |
| NUFIP1 | K-562 | ENCODE | Homo sapiens | ENCSR574XEO | Download (313) | ||
| NUP98-HOXA9 | HEK293-FT | GEO | Homo sapiens | GSE62586 | Download (36,576) | ||
| NUTM1 | NMC24335 | GEO | Homo sapiens | GSE96775 | Download (33,592) | ||
| OCA2 | HBL1 | GEO | Homo sapiens | GSE79480 | Download (163,403) | ||
| OGG1 | HEK293 | GEO | Homo sapiens | GSE89017 | Download (62,654) | ||
| OGG1 | HEK293 | 15min | GEO | Homo sapiens | GSE89017 | Download (39,751) | |
| OGG1 | HEK293 | 30_min | GEO | Homo sapiens | GSE89017 | Download (15,382) | |
| OGG1 | HEK293 | 60_min | GEO | Homo sapiens | GSE89017 | Download (32,185) | |
| ONECUT1 | liver | ENA | Homo sapiens | ERP002306 | Download (38,377) | ||
| ONECUT1 | H9 | ENA | Homo sapiens | ERP004206 | Download (50,558) | ||
| OSR2 | HEK293 | ENCODE | Homo sapiens | ENCSR324LTM | Download (43,580) | ||
| OSR2 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (30,235) | ||
| OTX2 | retina | pigment | GEO | Homo sapiens | GSE60024 | Download (15,952) | |
| OTX2 | D283-Med | GEO | Homo sapiens | GSE92582 | Download (119,339) | ||
| OTX2 | D283-Med | shGFP | GEO | Homo sapiens | GSE92582 | Download (154,444) | |
| OTX2 | D283-Med | shOTX2-1393 | GEO | Homo sapiens | GSE92582 | Download (123,308) | |
| OTX2 | D341-Med | GEO | Homo sapiens | GSE92582 | Download (99,716) | ||
| OTX2 | D341-Med | CRISPR-GFP1 | GEO | Homo sapiens | GSE92582 | Download (126,232) | |
| OTX2 | D341-Med | CRISPR-OTX2 | GEO | Homo sapiens | GSE92582 | Download (92,619) | |
| OTX2 | D341-Med | shGFP | GEO | Homo sapiens | GSE92582 | Download (115,909) | |
| OTX2 | D341-Med | shOTX2-1393 | GEO | Homo sapiens | GSE92582 | Download (102,573) | |
| OVOL2 | HCEC | GEO | Homo sapiens | GSE72641 | Download (13) | ||
| OVOL2 | HCET | GEO | Homo sapiens | GSE72641 | Download (4) | ||
| OVOL3 | HEK293 | ENCODE | Homo sapiens | ENCSR768LIO | Download (7,190) | ||
| PAF1 | HCT-116 | 37c | GEO | Homo sapiens | GSE97527 | Download (91,305) | |
| PAF1 | HCT-116 | ab20662 | GEO | Homo sapiens | GSE97527 | Download (61,556) | |
| PAF1 | HCT-116 | ab-sc-74797 | GEO | Homo sapiens | GSE97527 | Download (113,308) | |
| PALB2 | MCF-10A | GEO | Homo sapiens | GSE40591 | Download (2,336) | ||
| PATZ1 | HEK293 | ENCODE | Homo sapiens | ENCSR966ULI | Download (53,670) | ||
| PATZ1 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (4,458) | ||
| PAX3-FOXO1 | RH3 | GEO | Homo sapiens | GSE83726 | Download (14,556) | ||
| PAX3-FOXO1 | Hs-352-Sk | GEO | Homo sapiens | GSE83725 | Download (8,930) | ||
| PAX3-FOXO1 | Hs-352-Sk | PAX3-FOXO1-vector | GEO | Homo sapiens | GSE83725 | Download (19,428) | |
| PAX5 | DOHH2 | GEO | Homo sapiens | GSE69558 | Download (1,325) | ||
| PAX5 | OCI-Ly7 | GEO | Homo sapiens | GSE69558 | Download (6,539) | ||
| PAX5 | fetal | testis | GEO | Homo sapiens | GSE100639 | Download (75,907) | |
| PAX5 | GM12878 | ENCODE | Homo sapiens | ENCSR000BHD | Download (71,119) | ||
| PAX5 | GM12878 | ENCODE | Homo sapiens | ENCSR000BHJ | Download (65,539) | ||
| PAX5 | GM12891 | ENCODE | Homo sapiens | ENCSR000BJH | Download (21,246) | ||
| PAX5 | GM12892 | ENCODE | Homo sapiens | ENCSR000BJI | Download (30,353) | ||
| PAX6 | retina | pigment | GEO | Homo sapiens | GSE60024 | Download (9,667) | |
| PAX6 | EndoC-betaH2 | GEO | Homo sapiens | GSE87530 | Download (5,367) | ||
| PAX7 | H9 | DOX | GEO | Homo sapiens | GSE98976 | Download (21,809) | |
| PAX8 | GM12878 | ENCODE | Homo sapiens | ENCSR192AFN | Download (1,843) | ||
| PAX8 | K-562 | ENCODE | Homo sapiens | ENCSR274CGK | Download (2,087) | ||
| PAX8 | MCF-7 | ENCODE | Homo sapiens | ENCSR949IWY | Download (1,169) | ||
| PAX8 | FT194 | GEO | Homo sapiens | GSE79893 | Download (9) | ||
| PAX8 | FT246 | GEO | Homo sapiens | GSE79893 | Download (261) | ||
| PAX8 | FT33 | GEO | Homo sapiens | GSE79893 | Download (12) | ||
| PAX8 | JHOS4 | GEO | Homo sapiens | GSE79893 | Download (10) | ||
| PAX8 | Kuramochi | GEO | Homo sapiens | GSE79893 | Download (28) | ||
| PAX8 | OVSAHO | GEO | Homo sapiens | GSE79893 | Download (75,141) | ||
| PBX1 | RCH-ACV | GEO | Homo sapiens | GSE85988 | Download (33,611) | ||
| PBX2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR849DFF | Download (4,435) | ||
| PBX2 | K-562 | ENCODE | Homo sapiens | ENCSR263DFP | Download (34,816) | ||
| PBX2 | K-562 | ENCODE | Homo sapiens | ENCSR633EIC | Download (3,428) | ||
| PBX2 | K-562 | ENCODE | Homo sapiens | ENCSR979UVS | Download (145,149) | ||
| PBX3 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BVE | Download (92,078) | ||
| PBX3 | GM12878 | ENCODE | Homo sapiens | ENCSR000BGR | Download (46,464) | ||
| PBX3 | A-549 | ENCODE | Homo sapiens | ENCSR000BTN | Download (13,839) | ||
| PBX4 | PC-3 | Pbx4overexp | GEO | Homo sapiens | GSE97570 | Download (140,021) | |
| PC-MYB1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (4,066) |
| PC-MYB1 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (7,013) |
| PCBP1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR872EVQ | Download (31,610) | ||
| PCBP1 | K-562 | ENCODE | Homo sapiens | ENCSR052PTN | Download (19,438) | ||
| PCBP2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR945NSF | Download (20,965) | ||
| PCBP2 | K-562 | ENCODE | Homo sapiens | ENCSR603REQ | Download (2,951) | ||
| PCGF1 | WA01 | GEO | Homo sapiens | GSE104690 | Download (31,249) | ||
| PCGF2 | NT2-D1 | GEO | Homo sapiens | GSE101538 | Download (3,294) | ||
| PCGF2 | fibroblast | MET | GEO | Homo sapiens | GSE55605 | Download (7,233) | |
| PCGF2 | fibroblast | OHT | GEO | Homo sapiens | GSE55605 | Download (3,989) | |
| PDF2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (172) |
| PDX1 | hiPSC | derived_PPs | GEO | Homo sapiens | GSE106949 | Download (47,573) | |
| PDX1 | hESC | GEO | Homo sapiens | GSE58685 | Download (11,788) | ||
| PDX1 | islet | ENA | Homo sapiens | ERP001456 | Download (54,902) | ||
| PDX1 | islet | ENA | Homo sapiens | ERP004003 | Download (80,819) | ||
| PDX1 | H9 | ENA | Homo sapiens | ERP004206 | Download (2,634) | ||
| PGR | breast | tumor_Male_26 | GEO | Homo sapiens | GSE104399 | Download (54,695) | |
| PGR | breast | tumor_Male_28 | GEO | Homo sapiens | GSE104399 | Download (25,702) | |
| PGR | breast | tumor_Male_30 | GEO | Homo sapiens | GSE104399 | Download (22,479) | |
| PGR | AB32 | GEO | Homo sapiens | GSE31129 | Download (49,447) | ||
| PGR | breast | tumor-xenograft_1_E2_P4 | GEO | Homo sapiens | GSE93108 | Download (151) | |
| PGR | T-47D-A | E2_R5020 | GEO | Homo sapiens | GSE80358 | Download (14,550) | |
| PGR | T-47D-A | R5020 | GEO | Homo sapiens | GSE80358 | Download (6,602) | |
| PGR | T-47D | CR3flp_P4 | GEO | Homo sapiens | GSE99479 | Download (112,429) | |
| PGR | T-47D | CR3flp_veh | GEO | Homo sapiens | GSE99479 | Download (5,899) | |
| PGR | T-47D-B | E2_R5020 | GEO | Homo sapiens | GSE80358 | Download (3,164) | |
| PGR | T-47D-B | R5020 | GEO | Homo sapiens | GSE80358 | Download (3,360) | |
| PGR | hESC | GEO | Homo sapiens | GSE62475 | Download (3,996) | ||
| PGR | hESC | GEO | Homo sapiens | GSE62475 | Download (10,530) | ||
| PGR | hESC | GEO | Homo sapiens | GSE69539 | Download (8,691) | ||
| PGR | Leiomyoma | RU486 | GEO | Homo sapiens | GSE40724 | Download (26,649) | |
| PGR | HUVEC-C | GEO | Homo sapiens | GSE43786 | Download (10) | ||
| PGR | HUVEC-C | PR | GEO | Homo sapiens | GSE43786 | Download (29) | |
| PGR | HUVEC-C | PR_PROGESTERON | GEO | Homo sapiens | GSE43786 | Download (4,665) | |
| PGR | MCF-7 | PROG | GEO | Homo sapiens | GSE68355 | Download (73,610) | |
| PGR | MCF-7 | R5020 | GEO | Homo sapiens | GSE68355 | Download (52,356) | |
| PGR | T-47D | GEO | Homo sapiens | GSE31129 | Download (28,744) | ||
| PGR | T-47D | GEO | Homo sapiens | GSE40724 | Download (97,265) | ||
| PGR | T-47D | PROG | GEO | Homo sapiens | GSE68355 | Download (73,273) | |
| PGR | T-47D | R5020 | GEO | Homo sapiens | GSE68355 | Download (64,691) | |
| PGR | T-47D | E2 | GEO | Homo sapiens | GSE68356 | Download (122) | |
| PGR | T-47D | E2PG | GEO | Homo sapiens | GSE68356 | Download (12,514) | |
| PGR | T-47D | PG | GEO | Homo sapiens | GSE68356 | Download (12,293) | |
| PHB2 | K-562 | ENCODE | Homo sapiens | ENCSR924GXX | Download (6,269) | ||
| PHB2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR170DNV | Download (1,531) | ||
| PHF20 | K-562 | ENCODE | Homo sapiens | ENCSR594SMP | Download (9,626) | ||
| PHF21A | K-562 | ENCODE | Homo sapiens | ENCSR119VCX | Download (3,068) | ||
| PHF5A | Hep-G2 | ENCODE | Homo sapiens | ENCSR191TLD | Download (56,012) | ||
| PHF8 | A-549 | ENCODE | Homo sapiens | ENCSR541AOQ | Download (15,940) | ||
| PHF8 | K-562 | ENCODE | Homo sapiens | ENCSR000AQH | Download (25,094) | ||
| PHF8 | WA01 | ENCODE | Homo sapiens | ENCSR000ATK | Download (28,145) | ||
| PHF8 | HeLa | GEO | Homo sapiens | GSE20303 | Download (2,953) | ||
| PHF8 | HeLa | GEO | Homo sapiens | GSE22478 | Download (7,464) | ||
| PHIP | HCT-116 | ab833 | GEO | Homo sapiens | GSE101646 | Download (79,635) | |
| PHIP | HCT-116 | ab834 | GEO | Homo sapiens | GSE101646 | Download (34,192) | |
| PHIP | HCT-116 | BRWD2-KO_ab833 | GEO | Homo sapiens | GSE101646 | Download (1,280) | |
| PHIP | HCT-116 | MLL1-KO__ab833 | GEO | Homo sapiens | GSE101646 | Download (67,864) | |
| PHIP | HCT-116 | MLL4-SET-KO_ab833 | GEO | Homo sapiens | GSE101646 | Download (17,929) | |
| PHIP | HCT-116 | MLL4-SET-KO_ab834 | GEO | Homo sapiens | GSE101646 | Download (13,025) | |
| PHIP | HEK293 | ab833 | GEO | Homo sapiens | GSE101646 | Download (67,329) | |
| PHIP | HEK293 | flag_brwd2_phip | GEO | Homo sapiens | GSE101646 | Download (147,081) | |
| PHL1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,490) |
| PHL11 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,313) |
| PHL12 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (6,861) |
| PHL7 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (6,035) |
| PHOX2B | Kelly | GEO | Homo sapiens | GSE94822 | Download (163,879) | ||
| PHOX2B | SK-N-BE2-C | GEO | Homo sapiens | GSE94822 | Download (71,919) | ||
| PHOX2B | CLB-Ga | GEO | Homo sapiens | GSE90683 | Download (71,490) | ||
| PI | Ler | Ler_flower | 4w-dex | GEO | Arabidopsis thaliana | GSE38358 | Download (6,790) |
| PIAS1 | LNCaP | DHT | GEO | Homo sapiens | GSE83860 | Download (99,577) | |
| PIAS1 | LNCaP | DHT_TNFA | GEO | Homo sapiens | GSE83860 | Download (92,566) | |
| PIAS1 | LNCaP | DMSO | GEO | Homo sapiens | GSE83860 | Download (112,535) | |
| PIAS1 | LNCaP | TNFA | GEO | Homo sapiens | GSE83860 | Download (136,684) | |
| PIF1 | Col-0 | Col-0_seedling | 3d | GEO | Arabidopsis thaliana | GSE43283 | Download (3,728) |
| PIF3 | Col-0 | Col-0_seedling | 2d | GEO | Arabidopsis thaliana | GSE39215 | Download (1,731) |
| PIF4 | Col-0 | Col-0_seedling | 3d | GEO | Arabidopsis thaliana | GSE43284 | Download (1,532) |
| PIF4 | Col-0 | Col-0_seedling | 5d | GEO | Arabidopsis thaliana | GSE68193 | Download (13,644) |
| PIF4 | Col-0 | Col-0_seedling-etiolated | GEO | Arabidopsis thaliana | GSE35315 | Download (1,973) | |
| PIF5 | Col-0 | Col-0_seedling | 14d | GEO | Arabidopsis thaliana | GSE35059 | Download (2,391) |
| PIF5 | Col-0 | Col-0_seedling | 5d | GEO | Arabidopsis thaliana | GSE68193 | Download (7,021) |
| PITX3 | SH-SY5Y | GEO | Homo sapiens | GSE93275 | Download (54,472) | ||
| PKNOX1 | MCF-7 | ENCODE | Homo sapiens | ENCSR986XYK | Download (39,109) | ||
| PKNOX1 | GM12878 | ENCODE | Homo sapiens | ENCSR711XNY | Download (58,662) | ||
| PKNOX1 | HEK293T | ENCODE | Homo sapiens | ENCSR233FAG | Download (48,760) | ||
| PKNOX1 | K-562 | ENCODE | Homo sapiens | ENCSR115SMW | Download (57,611) | ||
| PLAG1 | K-562 | GEO | Homo sapiens | GSE111469 | Download (72,110) | ||
| PLRG1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR019KPC | Download (2,828) | ||
| PLT1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (245) |
| PML | GM12878 | ENCODE | Homo sapiens | ENCSR000BQM | Download (21,871) | ||
| PML | K-562 | ENCODE | Homo sapiens | ENCSR000BQY | Download (41,628) | ||
| PML | MCF-7 | ENCODE | Homo sapiens | ENCSR000BUZ | Download (13,577) | ||
| POU2F1 | IMR-90 | TERT | GEO | Homo sapiens | GSE38303 | Download (3,629) | |
| POU2F2 | HNPC | UNDIF | GEO | Homo sapiens | GSE74814 | Download (25,777) | |
| POU2F2 | BJAB | BIRA | GEO | Homo sapiens | GSE79480 | Download (769) | |
| POU2F2 | BJAB | BIRA_T223A | GEO | Homo sapiens | GSE79480 | Download (865) | |
| POU2F2 | HBL1 | GEO | Homo sapiens | GSE79480 | Download (176,068) | ||
| POU2F2 | GM12878 | ENCODE | Homo sapiens | ENCSR000BGP | Download (97,216) | ||
| POU2F2 | GM12891 | ENCODE | Homo sapiens | ENCSR000BII | Download (14,258) | ||
| POU2F3 | NCI-H1048 | GEO | Homo sapiens | GSE115123 | Download (66,579) | ||
| POU4F2 | HNPC | DIF | GEO | Homo sapiens | GSE74814 | Download (58,831) | |
| POU5F1 | hESC | GEO | Homo sapiens | GSE17917 | Download (89) | ||
| POU5F1 | hESC | GEO | Homo sapiens | GSE20650 | Download (268) | ||
| POU5F1 | hESC | NAIVE | GEO | Homo sapiens | GSE69646 | Download (76,522) | |
| POU5F1 | hESC | PRIMED | GEO | Homo sapiens | GSE69646 | Download (29,056) | |
| POU5F1 | NCCIT | GEO | Homo sapiens | GSE36134 | Download (5,455) | ||
| POU5F1 | NCCIT | SNF5 | GEO | Homo sapiens | GSE36134 | Download (36,360) | |
| POU5F1 | BJ1-hTERT | GEO | Homo sapiens | GSE92491 | Download (19,919) | ||
| POU5F1 | fetal | testis | GEO | Homo sapiens | GSE100639 | Download (118,928) | |
| POU5F1 | hESC | ENCODE | Homo sapiens | ENCSR264RJX | Download (10,900) | ||
| POU5F1 | WA09 | GEO | Homo sapiens | GSE105028 | Download (6,604) | ||
| POU5F1 | WA09 | heat-shock | GEO | Homo sapiens | GSE105028 | Download (5,242) | |
| POU5F1 | K-562 | ENCODE | Homo sapiens | ENCSR364SNE | Download (5,505) | ||
| POU5F1 | hiPSC | GEO | Homo sapiens | GSE56567 | Download (31,141) | ||
| POU5F1 | hiPSC | 3s2 | GEO | Homo sapiens | GSE81899 | Download (7,936) | |
| POU5F1 | OSK | GEO | Homo sapiens | GSE81899 | Download (5,954) | ||
| POU5F1 | OSKM | GEO | Homo sapiens | GSE81899 | Download (10,367) | ||
| POU5F1 | OSvK | GEO | Homo sapiens | GSE81899 | Download (682) | ||
| POU5F1 | OSvKM | GEO | Homo sapiens | GSE81899 | Download (1,502) | ||
| POU5F1 | WA01 | ENCODE | Homo sapiens | ENCSR000BMU | Download (4,512) | ||
| POU5F1 | WA01 | 3IL | ENA | Homo sapiens | ERP004238 | Download (32,792) | |
| POU5F1 | neuron | bipolar_doxy_4d | ENCODE | Homo sapiens | ENCSR362VCG | Download (3,232) | |
| POU5F1 | BGO3 | GEO | Homo sapiens | GSE21614 | Download (75,386) | ||
| POU5F1 | BJ | GEO | Homo sapiens | GSE36570 | Download (769) | ||
| POU5F1 | BJ | INDUCED | GEO | Homo sapiens | GSE36570 | Download (50,804) | |
| PPARG | WPMY-1 | ENA | Homo sapiens | ERP000333 | Download (567) | ||
| PPARG | ASC | GEO | Homo sapiens | GSE21366 | Download (24,901) | ||
| PPARG | macrophage | ENA | Homo sapiens | ERP008801 | Download (621) | ||
| PPARG | Hep-G2 | ENCODE | Homo sapiens | ENCSR130VQL | Download (79,137) | ||
| PPARG | HT29 | ROSIG_2H | GEO | Homo sapiens | GSE77039 | Download (26,553) | |
| PPARG | HT29 | ROSIG_48H | GEO | Homo sapiens | GSE77039 | Download (16,974) | |
| PPARG | SGBS | GEO | Homo sapiens | GSE41629 | Download (11,945) | ||
| PPARG | HUVEC-C | DMSO_HYPO | GEO | Homo sapiens | GSE50144 | Download (25,665) | |
| PPARG | HUVEC-C | DMSO_NORMO | GEO | Homo sapiens | GSE50144 | Download (536) | |
| PPARG | HUVEC-C | PPARG_HYPO | GEO | Homo sapiens | GSE50144 | Download (8,855) | |
| PPARG | HUVEC-C | PPARG_NORMO | GEO | Homo sapiens | GSE50144 | Download (189) | |
| PPARG | THP-1 | DIFF | GEO | Homo sapiens | GSE25426 | Download (1,039) | |
| PPARGC1A | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEQ | Download (170) | ||
| PRDM1 | fetal | testis | GEO | Homo sapiens | GSE100639 | Download (78,548) | |
| PRDM1 | HEK293 | ENCODE | Homo sapiens | ENCSR098YLE | Download (112,077) | ||
| PRDM1 | U266B1 | GEO | Homo sapiens | GSE102360 | Download (1,800) | ||
| PRDM1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECY | Download (4,564) | ||
| PRDM1 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (17,997) | ||
| PRDM10 | HEK293 | ENCODE | Homo sapiens | ENCSR223TAV | Download (96,830) | ||
| PRDM10 | K-562 | ENCODE | Homo sapiens | ENCSR120MPG | Download (26,434) | ||
| PRDM12 | HEK293 | ENCODE | Homo sapiens | ENCSR760AAY | Download (5,812) | ||
| PRDM14 | hESC | GEO | Homo sapiens | GSE22767 | Download (4,302) | ||
| PRDM14 | NCCIT | GEO | Homo sapiens | GSE71675 | Download (45,942) | ||
| PRDM2 | HEK293 | ENCODE | Homo sapiens | ENCSR714LYA | Download (772) | ||
| PRDM4 | HEK293 | ENCODE | Homo sapiens | ENCSR443MVV | Download (22,061) | ||
| PRDM6 | HEK293 | ENCODE | Homo sapiens | ENCSR892QHR | Download (77,320) | ||
| PRDM6 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (34,293) | ||
| PRDM9 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (103,095) | ||
| PRKDC | fibroblast | MET | GEO | Homo sapiens | GSE55605 | Download (689) | |
| PRKDC | fibroblast | OHT | GEO | Homo sapiens | GSE55605 | Download (1,481) | |
| PRKDC | MCF-7 | E2 | GEO | Homo sapiens | GSE60270 | Download (10,152) | |
| PRKDC | MCF-7 | SHCTR | GEO | Homo sapiens | GSE60270 | Download (21) | |
| PRKDC | MCF-7 | SHRARS | GEO | Homo sapiens | GSE60270 | Download (26) | |
| PRKDC | MCF-7 | SHRARS_E2 | GEO | Homo sapiens | GSE60270 | Download (17) | |
| PROX1 | SW480 | GEO | Homo sapiens | GSE60390 | Download (41,118) | ||
| PROX1 | HUVEC-C | Prox1OE | GEO | Homo sapiens | GSE71230 | Download (1,942) | |
| PRPF4 | Hep-G2 | ENCODE | Homo sapiens | ENCSR243LNQ | Download (22,704) | ||
| PRPF4 | K-562 | ENCODE | Homo sapiens | ENCSR220YXI | Download (7,445) | ||
| PSIP1 | ML-2 | GEO | Homo sapiens | GSE95511 | Download (1,379) | ||
| PTBP1 | K-562 | ENCODE | Homo sapiens | ENCSR948KMB | Download (16,096) | ||
| PTBP1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR156APP | Download (54,351) | ||
| PTRF | K-562 | ENCODE | Homo sapiens | ENCSR126FZN | Download (4,285) | ||
| PTTG1 | K-562 | ENCODE | Homo sapiens | ENCSR314BBS | Download (4,213) | ||
| PYGO2 | K-562 | ENCODE | Homo sapiens | ENCSR410DWC | Download (4,516) | ||
| PYGO2 | K-562 | ENCODE | Homo sapiens | ENCSR431XGJ | Download (2,624) | ||
| RAD21 | CHRF28811 | ENA | Homo sapiens | ERP008568 | Download (11,974) | ||
| RAD21 | GM12878 | ENCODE | Homo sapiens | ENCSR000BMY | Download (58,553) | ||
| RAD21 | GM12878 | ENCODE | Homo sapiens | ENCSR000EAC | Download (33,604) | ||
| RAD21 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BTU | Download (57,324) | ||
| RAD21 | liver | ENCODE | Homo sapiens | ENCSR230ZWH | Download (122,642) | ||
| RAD21 | liver | ENCODE | Homo sapiens | ENCSR635OSG | Download (20,423) | ||
| RAD21 | liver | ENCODE | Homo sapiens | ENCSR917QNE | Download (87,798) | ||
| RAD21 | neural | ENCODE | Homo sapiens | ENCSR198ZYJ | Download (47,883) | ||
| RAD21 | WA09 | GEO | Homo sapiens | GSE105028 | Download (46,244) | ||
| RAD21 | WA09 | heat-shock | GEO | Homo sapiens | GSE105028 | Download (41,795) | |
| RAD21 | A-549 | ENCODE | Homo sapiens | ENCSR000BUC | Download (55,370) | ||
| RAD21 | A-549 | ENCODE | Homo sapiens | ENCSR000DYE | Download (24,236) | ||
| RAD21 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BLS | Download (93,090) | ||
| RAD21 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEG | Download (55,049) | ||
| RAD21 | Hep-G2 | ENCODE | Homo sapiens | ENCSR054FKH | Download (58,928) | ||
| RAD21 | Hep-G2 | ENA | Homo sapiens | ERP000209 | Download (33,773) | ||
| RAD21 | IMR-90 | ENCODE | Homo sapiens | ENCSR000EFJ | Download (75,733) | ||
| RAD21 | K-562 | ENCODE | Homo sapiens | ENCSR000BKV | Download (63,527) | ||
| RAD21 | K-562 | ENCODE | Homo sapiens | ENCSR000FAD | Download (21,897) | ||
| RAD21 | HeLa-Tet-On | GEO | Homo sapiens | GSE112028 | Download (45,014) | ||
| RAD21 | HUVEC-C | hypoxia | GEO | Homo sapiens | GSE94872 | Download (65,753) | |
| RAD21 | HUVEC-C | normoxia | GEO | Homo sapiens | GSE94872 | Download (36,135) | |
| RAD21 | HUES-64 | GEO | Homo sapiens | GSE97394 | Download (92,587) | ||
| RAD21 | HUES-64 | DNMT-KO | GEO | Homo sapiens | GSE97394 | Download (130,087) | |
| RAD21 | LoVo | PHASEM | GEO | Homo sapiens | GSE51290 | Download (26,058) | |
| RAD21 | LoVo | PHASES | GEO | Homo sapiens | GSE51290 | Download (19,724) | |
| RAD21 | Hep-G2 | GEO | Homo sapiens | GSE72082 | Download (115,450) | ||
| RAD21 | IMR-5 | GEO | Homo sapiens | GSE78957 | Download (33,547) | ||
| RAD21 | MCF-7 | GEO | Homo sapiens | GSE72082 | Download (73,843) | ||
| RAD21 | RH4 | GEO | Homo sapiens | GSE83726 | Download (72,623) | ||
| RAD21 | SK-N-SH | GEO | Homo sapiens | GSE76815 | Download (21,117) | ||
| RAD21 | THP-1 | GEO | Homo sapiens | GSE55407 | Download (24,043) | ||
| RAD21 | MCF-7 | ENCODE | Homo sapiens | ENCSR000BTQ | Download (64,064) | ||
| RAD21 | MCF-7 | ENCODE | Homo sapiens | ENCSR703TNG | Download (75,230) | ||
| RAD21 | MCF-7 | ENA | Homo sapiens | ERP000209 | Download (62,678) | ||
| RAD21 | MCF-7 | E2 | ENA | Homo sapiens | ERP000209 | Download (52,644) | |
| RAD21 | MCF-7 | E2_SHCTCF | ENA | Homo sapiens | ERP000209 | Download (47,468) | |
| RAD21 | MCF-7 | E2_SHCTR | ENA | Homo sapiens | ERP000209 | Download (80,159) | |
| RAD21 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BLY | Download (104,287) | ||
| RAD21 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000EHX | Download (63,000) | ||
| RAD21 | THP-1 | eGFP-IFNb | GEO | Homo sapiens | GSE103477 | Download (76,885) | |
| RAD21 | THP-1 | NS1-IFNb | GEO | Homo sapiens | GSE103477 | Download (26,093) | |
| RAD21 | THP-1 | Pam3csk-000m-Flavo-000m | GEO | Homo sapiens | GSE103477 | Download (53,587) | |
| RAD21 | THP-1 | Pam3csk-000m-Flavo-240m | GEO | Homo sapiens | GSE103477 | Download (51,389) | |
| RAD21 | THP-1 | Pam3csk-020m-Flavo-000m | GEO | Homo sapiens | GSE103477 | Download (56,373) | |
| RAD21 | THP-1 | Pam3csk-025m-Flavo-000m | GEO | Homo sapiens | GSE103477 | Download (48,001) | |
| RAD21 | THP-1 | Pam3csk-030m-Flavo-000m | GEO | Homo sapiens | GSE103477 | Download (47,302) | |
| RAD21 | THP-1 | Pam3csk-045m-Flavo-000m | GEO | Homo sapiens | GSE103477 | Download (45,228) | |
| RAD21 | THP-1 | Pam3csk-060m-Flavo-000m | GEO | Homo sapiens | GSE103477 | Download (42,947) | |
| RAD21 | THP-1 | Pam3csk-120m-Flavo-000m | GEO | Homo sapiens | GSE103477 | Download (46,844) | |
| RAD21 | THP-1 | Pam3csk-150m-Flavo-030m | GEO | Homo sapiens | GSE103477 | Download (57,528) | |
| RAD21 | THP-1 | Pam3csk-180m-Flavo-060m | GEO | Homo sapiens | GSE103477 | Download (45,044) | |
| RAD21 | THP-1 | Pam3csk-360m-Flavo-240m | GEO | Homo sapiens | GSE103477 | Download (59,691) | |
| RAD21 | THP-1 | PMA_Dex-0h | GEO | Homo sapiens | GSE103477 | Download (39,054) | |
| RAD21 | THP-1 | PMA_Dex-6h | GEO | Homo sapiens | GSE103477 | Download (41,443) | |
| RAD21 | THP-1 | PMA_IFNb-0h | GEO | Homo sapiens | GSE103477 | Download (44,030) | |
| RAD21 | THP-1 | PMA_IFNb-6h | GEO | Homo sapiens | GSE103477 | Download (56,912) | |
| RAD21 | THP-1 | siCtrl-eGFP-Pam3csk-0h | GEO | Homo sapiens | GSE103477 | Download (27,533) | |
| RAD21 | THP-1 | siCtrl-eGFP-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (27,663) | |
| RAD21 | THP-1 | siCtrl-NS1-Pam3csk-0h | GEO | Homo sapiens | GSE103477 | Download (28,672) | |
| RAD21 | THP-1 | siCtrl-NS1-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (28,692) | |
| RAD21 | THP-1 | siCtrl-NS1-Pam3csk-7h-Flavo-3h | GEO | Homo sapiens | GSE103477 | Download (21,385) | |
| RAD21 | THP-1 | siNIPBL-eGFP-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (21,870) | |
| RAD21 | THP-1 | siNIPBL-NS1-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (17,844) | |
| RAD21 | THP-1 | siNIPBL-NS1-Pam3csk-7h-Flavo-3h | GEO | Homo sapiens | GSE103477 | Download (17,741) | |
| RAD21 | THP-1 | siWAPL-eGFP-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (28,782) | |
| RAD21 | THP-1 | siWAPL-NS1-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (26,078) | |
| RAD21 | THP-1 | siWAPL-NS1-Pam3csk-7h-Flavo-3h | GEO | Homo sapiens | GSE103477 | Download (22,535) | |
| RAD21 | WA01 | ENCODE | Homo sapiens | ENCSR000BLD | Download (102,513) | ||
| RAD21 | WA01 | ENCODE | Homo sapiens | ENCSR000ECE | Download (81,646) | ||
| RAD21 | HCT-116 | ENCODE | Homo sapiens | ENCSR000BSB | Download (53,460) | ||
| RAD21 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EDE | Download (82,323) | ||
| RAD21 | MDM | GEO | Homo sapiens | GSE103477 | Download (46,847) | ||
| RAD21 | MDM | -dNS1 | GEO | Homo sapiens | GSE103477 | Download (48,906) | |
| RAD21 | MDM | H5N1 | GEO | Homo sapiens | GSE103477 | Download (42,577) | |
| RAD21 | MDM | IFNb | GEO | Homo sapiens | GSE103477 | Download (43,370) | |
| RAD21 | HCT-116 | RAD21-mAC | GEO | Homo sapiens | GSE104888 | Download (41,921) | |
| RAD21 | HCT-116 | RAD21-mAC_500uM_auxin | GEO | Homo sapiens | GSE104888 | Download (6,434) | |
| RAD21 | GP5D | GEO | Homo sapiens | GSE51234 | Download (58,306) | ||
| RAD21 | GP5D | SIRAD21 | GEO | Homo sapiens | GSE51234 | Download (21,640) | |
| RAD51 | MCF-7 | ENCODE | Homo sapiens | ENCSR442VBJ | Download (8,114) | ||
| RAD51 | GM12878 | ENCODE | Homo sapiens | ENCSR482TWQ | Download (29,006) | ||
| RAD51 | U2OS | UASISIER | ENA | Homo sapiens | ERP001996 | Download (541) | |
| RAD51 | Hep-G2 | ENCODE | Homo sapiens | ENCSR081WLS | Download (6,286) | ||
| RAD51 | K-562 | ENCODE | Homo sapiens | ENCSR524BUE | Download (16,611) | ||
| RAD51 | U2OS | GEO | Homo sapiens | GSE90967 | Download (101) | ||
| RAD51 | U2OS | CX-5461 | GEO | Homo sapiens | GSE90967 | Download (1,100) | |
| RAP2-1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,158) |
| RAP2-10 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,941) |
| RAP2-11 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (839) |
| RAP2-12 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (766) |
| RAP2-6 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (858) |
| RARA | U-937 | ATRA-treated_RAR | GEO | Homo sapiens | GSE98006 | Download (2,875) | |
| RARA | SK-N-SH | GEO | Homo sapiens | GSE69119 | Download (1,841) | ||
| RARA | TSU-1621MT | GEO | Homo sapiens | GSE60477 | Download (25,471) | ||
| RARA | Hep-G2 | ENCODE | Homo sapiens | ENCSR500WXT | Download (107,923) | ||
| RAV1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,140) |
| RB1 | MCF-7 | siGFP | GEO | Homo sapiens | GSE98728 | Download (11,617) | |
| RB1 | MCF-7 | siH1 | GEO | Homo sapiens | GSE98728 | Download (9,551) | |
| RB1 | GM12878 | ENCODE | Homo sapiens | ENCSR785OKZ | Download (41,597) | ||
| RB1 | GM12878 | ENCODE | Homo sapiens | ENCSR916BOH | Download (2,969) | ||
| RB1 | K-562 | ENCODE | Homo sapiens | ENCSR506CVF | Download (19,876) | ||
| RB1 | K-562 | ENCODE | Homo sapiens | ENCSR670JDQ | Download (25,380) | ||
| RB1 | fibroblast | SENESCENT | GEO | Homo sapiens | GSE19898 | Download (104) | |
| RBAK | HEK293T | GEO | Homo sapiens | GSE78099 | Download (688) | ||
| RBAK | HEK293 | ENCODE | Homo sapiens | ENCSR441UBA | Download (2,272) | ||
| RBBP5 | GM12878 | ENCODE | Homo sapiens | ENCSR330EXS | Download (2,548) | ||
| RBBP5 | K-562 | ENCODE | Homo sapiens | ENCSR000AQI | Download (27,188) | ||
| RBBP5 | WA01 | ENCODE | Homo sapiens | ENCSR000AQC | Download (59,247) | ||
| RBFOX2 | K-562 | ENCODE | Homo sapiens | ENCSR822LBD | Download (63,975) | ||
| RBFOX2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR784FYS | Download (96,904) | ||
| RBM14 | K-562 | ENCODE | Homo sapiens | ENCSR423FCW | Download (8,396) | ||
| RBM15 | K-562 | ENCODE | Homo sapiens | ENCSR850WUE | Download (114) | ||
| RBM22 | K-562 | ENCODE | Homo sapiens | ENCSR848AOP | Download (59,059) | ||
| RBM22 | Hep-G2 | ENCODE | Homo sapiens | ENCSR356ECR | Download (266,587) | ||
| RBM25 | K-562 | ENCODE | Homo sapiens | ENCSR791OZM | Download (74,362) | ||
| RBM34 | K-562 | ENCODE | Homo sapiens | ENCSR899GSH | Download (466) | ||
| RBM39 | K-562 | ENCODE | Homo sapiens | ENCSR764OXF | Download (29,454) | ||
| RBM39 | Hep-G2 | ENCODE | Homo sapiens | ENCSR339JTP | Download (48,700) | ||
| RBOHJ | Col-0 | Col-0_seedling | 4d-DEX-6h | GEO | Arabidopsis thaliana | GSE80566 | Download (17,936) |
| RBP2 | U-937 | GEO | Homo sapiens | GSE28323 | Download (469) | ||
| RBP2 | U-937 | GEO | Homo sapiens | GSE28323 | Download (2,199) | ||
| RBPJ | LCL | GEO | Homo sapiens | GSE75503 | Download (25,233) | ||
| RBPJ | MUTUL | GEO | Homo sapiens | GSE75503 | Download (26,921) | ||
| RBPJ | NHEK | GEO | Homo sapiens | GSE29498 | Download (33,134) | ||
| RBPJ | GSC8-11 | GEO | Homo sapiens | GSE74557 | Download (4,005) | ||
| RBPJ | GSC8-11 | 12d-das | GEO | Homo sapiens | GSE74557 | Download (31,730) | |
| RBPJ | GSC8-11 | dasatinib | GEO | Homo sapiens | GSE74557 | Download (27,257) | |
| RBPJ | HUVEC-C | VEGF_12h | GEO | Homo sapiens | GSE109625 | Download (798) | |
| RBPJ | Hep-G2 | ENCODE | Homo sapiens | ENCSR596FEL | Download (84,885) | ||
| RBPJ | HKC | GEO | Homo sapiens | GSE102761 | Download (556) | ||
| RBPJ | CUTLL1 | GEO | Homo sapiens | GSE29600 | Download (10,499) | ||
| RBPJ | fibroblast | DERMAL | GEO | Homo sapiens | GSE59942 | Download (6) | |
| RBPJ | GIC | GEO | Homo sapiens | GSE79734 | Download (11,227) | ||
| RCOR1 | MCF-7 | ENCODE | Homo sapiens | ENCSR391JII | Download (9,764) | ||
| RCOR1 | SK-N-SH | ENCODE | Homo sapiens | ENCSR009TKN | Download (72,684) | ||
| RCOR1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECM | Download (27,184) | ||
| RCOR1 | HeLa | GEO | Homo sapiens | GSE45441 | Download (11,169) | ||
| RCOR1 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZC | Download (489) | ||
| RCOR1 | A-549 | ENCODE | Homo sapiens | ENCSR618ICR | Download (2,362) | ||
| RCOR1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EDQ | Download (16,995) | ||
| RCOR1 | IMR-90 | ENCODE | Homo sapiens | ENCSR000EFG | Download (43,574) | ||
| RCOR1 | K-562 | ENCODE | Homo sapiens | ENCSR000EGC | Download (16,119) | ||
| RCOR1 | K-562 | ENCODE | Homo sapiens | ENCSR000EGG | Download (108,206) | ||
| RCOR1 | keratinocyte | diff | GEO | Homo sapiens | GSE57702 | Download (70,005) | |
| RCOR1 | AML | GEO | Homo sapiens | GSE112074 | Download (9,567) | ||
| RCOR1 | AML | OG86 | GEO | Homo sapiens | GSE112074 | Download (10,145) | |
| RCOR2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR455DOO | Download (80,428) | ||
| RD26 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (21,409) |
| RD26 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (19,708) |
| RELA | HeLa-B2 | DMSO | GEO | Homo sapiens | GSE24518 | Download (3,158) | |
| RELA | HeLa-B2 | GRKD_DMSO | GEO | Homo sapiens | GSE24518 | Download (12,452) | |
| RELA | HeLa-B2 | GRKD_TA_TNFA | GEO | Homo sapiens | GSE24518 | Download (10,511) | |
| RELA | HeLa-B2 | P65KD_DMSO | GEO | Homo sapiens | GSE24518 | Download (7,393) | |
| RELA | HeLa-B2 | P65KD_TA_TNFA | GEO | Homo sapiens | GSE24518 | Download (8,413) | |
| RELA | HeLa-B2 | TA | GEO | Homo sapiens | GSE24518 | Download (1,358) | |
| RELA | HeLa-B2 | TA_TNFA | GEO | Homo sapiens | GSE24518 | Download (18,284) | |
| RELA | HeLa-B2 | TNFA | GEO | Homo sapiens | GSE24518 | Download (21,829) | |
| RELA | HAEC | GEO | Homo sapiens | GSE89970 | Download (31,745) | ||
| RELA | HAEC | IL1b_4h | GEO | Homo sapiens | GSE89970 | Download (58,062) | |
| RELA | HAEC | TNFa_4h | GEO | Homo sapiens | GSE89970 | Download (77,150) | |
| RELA | Detroit-562 | LPS | GEO | Homo sapiens | GSE91018 | Download (29,303) | |
| RELA | Detroit-562 | Pam2CSK4 | GEO | Homo sapiens | GSE91018 | Download (24,053) | |
| RELA | Detroit-562 | Poly-I | GEO | Homo sapiens | GSE91018 | Download (40,025) | |
| RELA | Detroit-562 | TNFa | GEO | Homo sapiens | GSE91018 | Download (38,351) | |
| RELA | Detroit-562 | tri-DAP | GEO | Homo sapiens | GSE91018 | Download (23,352) | |
| RELA | 786-M1A | GEO | Homo sapiens | GSE98012 | Download (14,239) | ||
| RELA | GM10847 | ENCODE | Homo sapiens | ENCSR000DYM | Download (6,633) | ||
| RELA | GM12878 | ENCODE | Homo sapiens | ENCSR000EAG | Download (24,520) | ||
| RELA | GM12878 | ENCODE | Homo sapiens | ENCSR664POU | Download (24,879) | ||
| RELA | GM12891 | ENCODE | Homo sapiens | ENCSR000EAI | Download (22,513) | ||
| RELA | GM12892 | ENCODE | Homo sapiens | ENCSR000EAN | Download (3,924) | ||
| RELA | GM15510 | ENCODE | Homo sapiens | ENCSR000EAQ | Download (8,989) | ||
| RELA | GM18505 | ENCODE | Homo sapiens | ENCSR000EAW | Download (7,025) | ||
| RELA | GM18526 | ENCODE | Homo sapiens | ENCSR000EBA | Download (7,344) | ||
| RELA | GM18951 | ENCODE | Homo sapiens | ENCSR000EBD | Download (6,538) | ||
| RELA | GM19099 | ENCODE | Homo sapiens | ENCSR000EBI | Download (24,649) | ||
| RELA | GM19193 | ENCODE | Homo sapiens | ENCSR000EBM | Download (4,814) | ||
| RELA | monocyte | nopretreatment | GEO | Homo sapiens | GSE100381 | Download (147,036) | |
| RELA | monocyte | notreatment | GEO | Homo sapiens | GSE100381 | Download (475,795) | |
| RELA | SW480 | 0h_TNFa | GEO | Homo sapiens | GSE102796 | Download (3,393) | |
| RELA | SW480 | 16h_TNFa | GEO | Homo sapiens | GSE102796 | Download (37,386) | |
| RELA | K-562 | ENCODE | Homo sapiens | ENCSR772EEN | Download (11,999) | ||
| RELA | THP-1 | eGFP-Pam3csk-0h | GEO | Homo sapiens | GSE103477 | Download (1,625) | |
| RELA | THP-1 | eGFP-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (19,408) | |
| RELA | AC16 | GEO | Homo sapiens | GSE51169 | Download (1,227) | ||
| RELA | AC16 | TNFA | GEO | Homo sapiens | GSE51169 | Download (6,816) | |
| RELA | BJAB | GEO | Homo sapiens | GSE117250 | Download (15,432) | ||
| RELA | BJAB | 1h-activation | GEO | Homo sapiens | GSE117250 | Download (22,076) | |
| RELA | BJAB | 4h-activation | GEO | Homo sapiens | GSE117250 | Download (11,374) | |
| RELA | CD4 | TH1_BAY | GEO | Homo sapiens | GSE62482 | Download (18,330) | |
| RELA | CD4 | TH1_DMSO | GEO | Homo sapiens | GSE62482 | Download (21,695) | |
| RELA | HDF | DMSO | GEO | Homo sapiens | GSE77225 | Download (4,573) | |
| RELA | HDF | NUTLIN | GEO | Homo sapiens | GSE77225 | Download (10,669) | |
| RELA | HEK293 | TNF-15min | GEO | Homo sapiens | GSE75562 | Download (573) | |
| RELA | HEK293 | TNF-1h | GEO | Homo sapiens | GSE75562 | Download (27,285) | |
| RELA | HEK293 | TNF-30min | GEO | Homo sapiens | GSE75562 | Download (2,720) | |
| RELA | HEK293 | GEO | Homo sapiens | GSE89017 | Download (1,170) | ||
| RELA | HEK293 | 15min | GEO | Homo sapiens | GSE89017 | Download (260) | |
| RELA | HEK293 | 30_min | GEO | Homo sapiens | GSE89017 | Download (2,134) | |
| RELA | HEK293 | 60_min | GEO | Homo sapiens | GSE89017 | Download (692) | |
| RELA | KB | GEO | Homo sapiens | GSE52469 | Download (18,957) | ||
| RELA | KB | IL | GEO | Homo sapiens | GSE52469 | Download (25,747) | |
| RELA | KB | GEO | Homo sapiens | GSE64223 | Download (1) | ||
| RELA | KB | 5Z | GEO | Homo sapiens | GSE64223 | Download (4,394) | |
| RELA | KB | 5Z_IL | GEO | Homo sapiens | GSE64223 | Download (3,943) | |
| RELA | KB | IL | GEO | Homo sapiens | GSE64223 | Download (2) | |
| RELA | KB | PHA | GEO | Homo sapiens | GSE64223 | Download (1) | |
| RELA | KB | PHA_IL | GEO | Homo sapiens | GSE64223 | Download (1) | |
| RELA | L1236 | GEO | Homo sapiens | GSE63736 | Download (612) | ||
| RELA | LNCaP | DHT_TNFA | GEO | Homo sapiens | GSE83860 | Download (20,451) | |
| RELA | LNCaP | SICTR_TNFA | GEO | Homo sapiens | GSE83860 | Download (29,236) | |
| RELA | LNCaP | SIFOXA1_TNFA | GEO | Homo sapiens | GSE83860 | Download (37,589) | |
| RELA | LNCaP | TNFA | GEO | Homo sapiens | GSE83860 | Download (2,573) | |
| RELA | SGBS | GEO | Homo sapiens | GSE64233 | Download (23,174) | ||
| RELA | SGBS | TNF | GEO | Homo sapiens | GSE64233 | Download (75,034) | |
| RELA | U2OS | GEO | Homo sapiens | GSE109996 | Download (2,267) | ||
| RELA | U2OS | 10Gy_2h_recovery | GEO | Homo sapiens | GSE109996 | Download (1,193) | |
| RELA | 786-O | GEO | Homo sapiens | GSE109953 | Download (25,140) | ||
| RELA | 786-O | GEO | Homo sapiens | GSE86092 | Download (69,699) | ||
| RELA | Huh-7 | IL1 | GEO | Homo sapiens | GSE89212 | Download (2,078) | |
| RELA | HUVEC-C | TNF_0M | GEO | Homo sapiens | GSE34500 | Download (901) | |
| RELA | HUVEC-C | TNF_30M | GEO | Homo sapiens | GSE34500 | Download (29,815) | |
| RELA | HUVEC-C | TNF | GEO | Homo sapiens | GSE43070 | Download (7,571) | |
| RELA | HUVEC-C | GEO | Homo sapiens | GSE53998 | Download (295) | ||
| RELA | HUVEC-C | TNF | GEO | Homo sapiens | GSE53998 | Download (16,746) | |
| RELA | HUVEC-C | TNF_JQ1 | GEO | Homo sapiens | GSE53998 | Download (19,548) | |
| RELA | HUVEC-C | Notch1kd_IL1beta | GEO | Homo sapiens | GSE87552 | Download (86,678) | |
| RELA | HUVEC-C | Scr | GEO | Homo sapiens | GSE87552 | Download (7,123) | |
| RELA | HUVEC-C | Scr_IL1beta | GEO | Homo sapiens | GSE87552 | Download (81,614) | |
| RELA | IMR-90 | GEO | Homo sapiens | GSE43070 | Download (803) | ||
| RELA | IMR-90 | TNF | GEO | Homo sapiens | GSE43070 | Download (22,640) | |
| RELA | MCF-7 | GEO | Homo sapiens | GSE59530 | Download (790) | ||
| RELA | MCF-7 | E2 | GEO | Homo sapiens | GSE59530 | Download (941) | |
| RELA | MCF-7 | E2_TNF | GEO | Homo sapiens | GSE59530 | Download (2,681) | |
| RELA | MCF-7 | TNF | GEO | Homo sapiens | GSE59530 | Download (1,572) | |
| RELA | MCF-7 | E2_45m | GEO | Homo sapiens | GSE67295 | Download (2,932) | |
| RELA | MCF-7 | IL1b_45m | GEO | Homo sapiens | GSE67295 | Download (27,098) | |
| RELA | MCF-7 | TNFa_45m | GEO | Homo sapiens | GSE67295 | Download (14,562) | |
| RELA | MCF-7 | Veh | GEO | Homo sapiens | GSE67295 | Download (27,874) | |
| RELB | L1236 | GEO | Homo sapiens | GSE63736 | Download (16,935) | ||
| RELB | GM12878 | ENCODE | Homo sapiens | ENCSR387QUV | Download (75,569) | ||
| REPIN1 | HEK293 | ENCODE | Homo sapiens | ENCSR146NLL | Download (5,507) | ||
| REST | GM12878 | ENCODE | Homo sapiens | ENCSR000BGF | Download (8,691) | ||
| REST | GM12878 | ENCODE | Homo sapiens | ENCSR000BQS | Download (20,703) | ||
| REST | HEK293 | ENCODE | Homo sapiens | ENCSR896UBV | Download (27,224) | ||
| REST | hepatocyte | ENA | Homo sapiens | ERP000395 | Download (1,186) | ||
| REST | Ishikawa | ENCODE | Homo sapiens | ENCSR000BUU | Download (13,561) | ||
| REST | liver | ENCODE | Homo sapiens | ENCSR867WPH | Download (41,981) | ||
| REST | liver | ENCODE | Homo sapiens | ENCSR893QWP | Download (27,993) | ||
| REST | neural | ENCODE | Homo sapiens | ENCSR000BTV | Download (57,836) | ||
| REST | PFSK1 | ENCODE | Homo sapiens | ENCSR000BJP | Download (7,954) | ||
| REST | PFSK1 | ENCODE | Homo sapiens | ENCSR000BOX | Download (20,958) | ||
| REST | A-549 | ENCODE | Homo sapiens | ENCSR000BQP | Download (49,377) | ||
| REST | A-549 | ENCODE | Homo sapiens | ENCSR892DRK | Download (12,543) | ||
| REST | GM23338 | ENCODE | Homo sapiens | ENCSR871KYB | Download (8,114) | ||
| REST | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BJL | Download (5,384) | ||
| REST | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BOT | Download (27,622) | ||
| REST | HL-60 | ENCODE | Homo sapiens | ENCSR000BTF | Download (11,250) | ||
| REST | K-562 | ENCODE | Homo sapiens | ENCSR000ATM | Download (7,923) | ||
| REST | K-562 | ENCODE | Homo sapiens | ENCSR000BMW | Download (54,019) | ||
| REST | K-562 | ENCODE | Homo sapiens | ENCSR137ZMQ | Download (53,595) | ||
| REST | K-562 | GEO | Homo sapiens | GSE70482 | Download (8,834) | ||
| REST | NCI-H295R | GEO | Homo sapiens | GSE49014 | Download (2,108) | ||
| REST | NCI-H295R | SF1 | GEO | Homo sapiens | GSE49014 | Download (2,021) | |
| REST | MCF-7 | ENCODE | Homo sapiens | ENCSR000BSP | Download (14,982) | ||
| REST | PANC-1 | ENCODE | Homo sapiens | ENCSR000BJO | Download (29,831) | ||
| REST | PANC-1 | ENCODE | Homo sapiens | ENCSR000BPK | Download (19,227) | ||
| REST | PANC-1 | ENCODE | Homo sapiens | ENCSR000BUP | Download (5,012) | ||
| REST | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BJJ | Download (7,774) | ||
| REST | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BOZ | Download (82,825) | ||
| REST | WA01 | ENCODE | Homo sapiens | ENCSR000BHM | Download (25,812) | ||
| REST | WA01 | ENCODE | Homo sapiens | ENCSR663WAR | Download (4,852) | ||
| REST | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000BMN | Download (23,160) | ||
| REST | HCT-116 | ENCODE | Homo sapiens | ENCSR000BVI | Download (4,429) | ||
| REST | CD4 | GEO | Homo sapiens | GSE49570 | Download (15,960) | ||
| REST | GP5D | GEO | Homo sapiens | GSE51234 | Download (3,098) | ||
| REST | GP5D | SIRAD21 | GEO | Homo sapiens | GSE51234 | Download (2,793) | |
| REV | Col-0 | Col-0_seedling | 10d-DEX-90min | GEO | Arabidopsis thaliana | GSE26722 | Download (8,497) |
| RFX1 | K-562 | ENCODE | Homo sapiens | ENCSR968GIB | Download (22,420) | ||
| RFX1 | MCF-7 | ENCODE | Homo sapiens | ENCSR066TET | Download (28,829) | ||
| RFX1 | MCF-7 | ENCODE | Homo sapiens | ENCSR788XNX | Download (17,784) | ||
| RFX1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR928API | Download (3,523) | ||
| RFX1 | K-562 | ENCODE | Homo sapiens | ENCSR041AXL | Download (17,904) | ||
| RFX2 | GP5D | GEO | Homo sapiens | GSE51234 | Download (870) | ||
| RFX2 | GP5D | SIRAD21 | GEO | Homo sapiens | GSE51234 | Download (58) | |
| RFX3 | Hep-G2 | ENCODE | Homo sapiens | ENCSR633OVO | Download (18,702) | ||
| RFX5 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZW | Download (1,151) | ||
| RFX5 | A-549 | ENCODE | Homo sapiens | ENCSR064LJN | Download (5,152) | ||
| RFX5 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEA | Download (14,171) | ||
| RFX5 | IMR-90 | ENCODE | Homo sapiens | ENCSR000EFD | Download (5,855) | ||
| RFX5 | K-562 | ENCODE | Homo sapiens | ENCSR000EGO | Download (2,334) | ||
| RFX5 | MCF-7 | ENCODE | Homo sapiens | ENCSR924TVL | Download (10,185) | ||
| RFX5 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000EHY | Download (6,132) | ||
| RFX5 | WA01 | ENCODE | Homo sapiens | ENCSR000ECF | Download (1,883) | ||
| RFX5 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECX | Download (32,501) | ||
| RGA | Ler | Ler_seedling | 11d | GEO | Arabidopsis thaliana | GSE59187 | Download (4,366) |
| RGA | Ler | Ler_inflorescence-apices | GEO | Arabidopsis thaliana | GSE94926 | Download (10,497) | |
| RING1B | WA09 | GEO | Homo sapiens | GSE105028 | Download (15,001) | ||
| RING1B | WA09 | heat-shock | GEO | Homo sapiens | GSE105028 | Download (14,361) | |
| RKD2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (549) |
| RLF | K-562 | ENCODE | Homo sapiens | ENCSR718SDE | Download (4,145) | ||
| RNF2 | K-562 | ENCODE | Homo sapiens | ENCSR608XTF | Download (84,082) | ||
| RNF2 | K-562 | ENCODE | Homo sapiens | ENCSR820GND | Download (35,078) | ||
| RNF2 | WA01 | ENCODE | Homo sapiens | ENCSR784VUY | Download (11,321) | ||
| RNF2 | WA01 | GEO | Homo sapiens | GSE104690 | Download (7,938) | ||
| RNF2 | HUES-64 | GEO | Homo sapiens | GSE104059 | Download (4,508) | ||
| RNF2 | HEK293T | GEO | Homo sapiens | GSE34774 | Download (17) | ||
| RNF2 | GM12878 | ENCODE | Homo sapiens | ENCSR679FAB | Download (23,125) | ||
| RNF2 | A-549 | ENCODE | Homo sapiens | ENCSR798EGJ | Download (23,457) | ||
| RNF2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR523XCR | Download (14,315) | ||
| RNF2 | K-562 | ENCODE | Homo sapiens | ENCSR076YPO | Download (24,066) | ||
| RNF2 | K-562 | ENCODE | Homo sapiens | ENCSR138FUZ | Download (30,144) | ||
| RNF2 | hESC | GEO | Homo sapiens | GSE13084 | Download (18) | ||
| RNF2 | HMELBRAF | OVER | GEO | Homo sapiens | GSE51929 | Download (13,470) | |
| RNF2 | HMELBRAF | OVERTUMOR | GEO | Homo sapiens | GSE51929 | Download (843) | |
| RNF2 | NCCIT | GEO | Homo sapiens | GSE71675 | Download (675) | ||
| RPL | Ler-0 | Ler-0_inflorescence-apices | GEO | Arabidopsis thaliana | GSE78727 | Download (11,583) | |
| RPS4 | Col-0 | Col-0_leaves | 3-5w-eds1 | GEO | Arabidopsis thaliana | GSE40329 | Download (2,679) |
| RPS4 | Col-0 | Col-0_leaves | 3-5w | GEO | Arabidopsis thaliana | GSE40329 | Download (2,344) |
| RUNX1 | ME-1 | GEO | Homo sapiens | GSE46044 | Download (34,861) | ||
| RUNX1 | Jurkat | GEO | Homo sapiens | GSE29180 | Download (10,041) | ||
| RUNX1 | Jurkat | GEO | Homo sapiens | GSE42575 | Download (5,020) | ||
| RUNX1 | Jurkat | GEO | Homo sapiens | GSE68976 | Download (35,003) | ||
| RUNX1 | Jurkat | GEO | Homo sapiens | GSE76181 | Download (44,066) | ||
| RUNX1 | Jurkat | GEO | Homo sapiens | GSE85524 | Download (39,533) | ||
| RUNX1 | NB4 | GEO | Homo sapiens | GSE81992 | Download (63,257) | ||
| RUNX1 | PBMC | GEO | Homo sapiens | GSE85524 | Download (103) | ||
| RUNX1 | SKNO1 | GEO | Homo sapiens | GSE23730 | Download (54,718) | ||
| RUNX1 | VCaP | DHT24H | GEO | Homo sapiens | GSE58428 | Download (61,063) | |
| RUNX1 | CCRF-CEM | GEO | Homo sapiens | GSE33850 | Download (620) | ||
| RUNX1 | Kasumi-1 | GEO | Homo sapiens | GSE45738 | Download (25,195) | ||
| RUNX1 | Kasumi-1 | GEO | Homo sapiens | GSE62847 | Download (21,154) | ||
| RUNX1 | Kasumi-1 | GEO | Homo sapiens | GSE65427 | Download (17,323) | ||
| RUNX1 | MCF-7 | GEO | Homo sapiens | GSE75070 | Download (181) | ||
| RUNX1 | MV4-11 | GEO | Homo sapiens | GSE79899 | Download (53,564) | ||
| RUNX1 | NALM-6 | GEO | Homo sapiens | GSE109377 | Download (3,652) | ||
| RUNX1 | SKH1 | GEO | Homo sapiens | GSE87283 | Download (37,554) | ||
| RUNX1 | SKH1 | 10d | GEO | Homo sapiens | GSE87283 | Download (48,651) | |
| RUNX1 | SKH1 | RUNX1-EVI1_KD | GEO | Homo sapiens | GSE87283 | Download (53,369) | |
| RUNX1 | THP-1 | GEO | Homo sapiens | GSE79899 | Download (37,528) | ||
| RUNX1 | TSU-1621MT | GEO | Homo sapiens | GSE60477 | Download (86,170) | ||
| RUNX1 | U-937 | GEO | Homo sapiens | GSE65427 | Download (9,411) | ||
| RUNX1 | BCP-ALL | patient1 | GEO | Homo sapiens | GSE109377 | Download (40,144) | |
| RUNX1 | K-562 | ENCODE | Homo sapiens | ENCSR588AKU | Download (13,882) | ||
| RUNX1 | ME-1 | Human-leukemia_AI-10-49 | GEO | Homo sapiens | GSE101789 | Download (10,229) | |
| RUNX1 | epididymis | HEE | GEO | Homo sapiens | GSE109061 | Download (56,788) | |
| RUNX1 | epididymis | HEE_R1881 | GEO | Homo sapiens | GSE109061 | Download (2,655) | |
| RUNX1 | AMLBLAST | GEO | Homo sapiens | GSE60130 | Download (47,132) | ||
| RUNX1 | BONE | MARROW | GEO | Homo sapiens | GSE64862 | Download (87,475) | |
| RUNX1 | CD34 | GEO | Homo sapiens | GSE64862 | Download (47,141) | ||
| RUNX1 | CD34 | ADULT | GEO | Homo sapiens | GSE70660 | Download (13,533) | |
| RUNX1 | CD34 | FETAL | GEO | Homo sapiens | GSE70660 | Download (6,292) | |
| RUNX1 | HEK293T | GEO | Homo sapiens | GSE85524 | Download (110,682) | ||
| RUNX1 | ALL-SIL | GEO | Homo sapiens | GSE102209 | Download (12,930) | ||
| RUNX1 | HL-60 | ENA | Homo sapiens | ERP008568 | Download (42) | ||
| RUNX1 | HL-60 | GEO | Homo sapiens | GSE107553 | Download (10,001) | ||
| RUNX1 | K-562 | ENCODE | Homo sapiens | ENCSR414TYY | Download (3,367) | ||
| RUNX1-3 | Jurkat | GEO | Homo sapiens | GSE17954 | Download (1,179) | ||
| RUNX1T1 | Kasumi-1 | GEO | Homo sapiens | GSE43834 | Download (28,880) | ||
| RUNX1T1 | Kasumi-1 | GEO | Homo sapiens | GSE65427 | Download (18,705) | ||
| RUNX1T1 | Kasumi-1 | GEO | Homo sapiens | GSE102697 | Download (46,025) | ||
| RUNX1T1 | Kasumi-1 | CEBPA-ER | GEO | Homo sapiens | GSE102697 | Download (27,679) | |
| RUNX1T1 | Kasumi-1 | CEBPA-ER_E2 | GEO | Homo sapiens | GSE102697 | Download (32,864) | |
| RUNX1T1 | Kasumi-1 | E2 | GEO | Homo sapiens | GSE102697 | Download (26,813) | |
| RUNX1T1 | CD34 | GEO | Homo sapiens | GSE80773 | Download (34,410) | ||
| RUNX2 | LNCaP-C4-2B | GEO | Homo sapiens | GSE33889 | Download (39,808) | ||
| RUNX2 | IMSC3 | ENA | Homo sapiens | ERP003787 | Download (257,600) | ||
| RUNX2 | SaOS-2 | GEO | Homo sapiens | GSE76937 | Download (6,452) | ||
| RUNX3 | GM12878 | ENCODE | Homo sapiens | ENCSR000BRI | Download (114,798) | ||
| RUVBL1 | Hep-G2 | GEO | Homo sapiens | GSE107730 | Download (1,048) | ||
| RUVBL1 | Hep-G2 | GEO | Homo sapiens | GSE97661 | Download (6,060) | ||
| RUVBL2 | Hep-G2 | GEO | Homo sapiens | GSE107730 | Download (5,920) | ||
| RVE4 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (523) |
| RVE5 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,118) |
| RVE6 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,407) |
| RVE7 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,540) |
| RXR | LS180 | GEO | Homo sapiens | GSE31939 | Download (11,894) | ||
| RXR | LS180 | 125 | GEO | Homo sapiens | GSE31939 | Download (12,089) | |
| RXR | macrophage | ENA | Homo sapiens | ERP008801 | Download (23,629) | ||
| RXR | macrophage | ENA | Homo sapiens | ERP009021 | Download (2,210) | ||
| RXR | THP-1 | DIFF | GEO | Homo sapiens | GSE25426 | Download (550) | |
| RXR | TSU-1621MT | GEO | Homo sapiens | GSE60477 | Download (40,335) | ||
| RXRA | GM12878 | ENCODE | Homo sapiens | ENCSR000BJD | Download (15,366) | ||
| RXRA | liver | ENCODE | Homo sapiens | ENCSR098XMN | Download (102,767) | ||
| RXRA | liver | ENCODE | Homo sapiens | ENCSR352QSB | Download (92,826) | ||
| RXRA | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BHU | Download (29,254) | ||
| RXRA | JMSU-1 | GEO | Homo sapiens | GSE107734 | Download (15,403) | ||
| RXRA | JMSU-1 | RXRA-S427F | GEO | Homo sapiens | GSE107734 | Download (15,452) | |
| RXRA | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BVG | Download (17,980) | ||
| RXRA | WA01 | ENCODE | Homo sapiens | ENCSR000BJW | Download (4,765) | ||
| RXRB | Hep-G2 | ENCODE | Homo sapiens | ENCSR560SEP | Download (103,221) | ||
| RYBP | WA01 | GEO | Homo sapiens | GSE104690 | Download (35,584) | ||
| RYBP | HEK293T | GEO | Homo sapiens | GSE34774 | Download (1,570) | ||
| S1FA3 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,422) |
| SAFB | K-562 | ENCODE | Homo sapiens | ENCSR072VUO | Download (6,336) | ||
| SALL1 | HEK293 | ENCODE | Homo sapiens | ENCSR041ZIQ | Download (551) | ||
| SALL2 | HEK293 | ENCODE | Homo sapiens | ENCSR044FMM | Download (1,809) | ||
| SALL4 | SNU-398 | GEO | Homo sapiens | GSE112729 | Download (1,185) | ||
| SAP130 | Hep-G2 | ENCODE | Homo sapiens | ENCSR959OMS | Download (106,172) | ||
| SAP30 | K-562 | ENCODE | Homo sapiens | ENCSR000AQJ | Download (17,025) | ||
| SAP30 | WA01 | ENCODE | Homo sapiens | ENCSR000ATR | Download (32,809) | ||
| SATB1 | Hep-G2 | GEO | Homo sapiens | GSE108514 | Download (2,419) | ||
| SATB1 | SH-SY5Y | GEO | Homo sapiens | GSE73576 | Download (893) | ||
| SATB1 | SH-SY5Y | ABETA | GEO | Homo sapiens | GSE73576 | Download (585) | |
| SCRT1 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (107,863) | ||
| SCRT1 | HEK293 | ENCODE | Homo sapiens | ENCSR605MGM | Download (34,502) | ||
| SCRT2 | HEK293 | ENCODE | Homo sapiens | ENCSR338DGO | Download (111,984) | ||
| SEP3 | Col-0 | Col-0_inflorescence | 5-7w | GEO | Arabidopsis thaliana | GSE14600 | Download (6,099) |
| SEP3 | Ler | Ler_inflorescence | 5-7w | GEO | Arabidopsis thaliana | GSE14600 | Download (5,862) |
| SEP3 | Ler | Ler_inflorescence | 4w-2d | GEO | Arabidopsis thaliana | GSE46986 | Download (4,306) |
| SEP3 | Ler | Ler_inflorescence | 4w-4d | GEO | Arabidopsis thaliana | GSE46986 | Download (9,530) |
| SEP3 | Ler | Ler_inflorescence | 4w-8d | GEO | Arabidopsis thaliana | GSE46986 | Download (10,211) |
| SETDB1 | HeLa | MORC2-KO | GEO | Homo sapiens | GSE95456 | Download (170,349) | |
| SETDB1 | HEK293 | ENCODE | Homo sapiens | ENCSR348AGV | Download (58,867) | ||
| SETDB1 | U2OS | ENCODE | Homo sapiens | ENCSR000EYD | Download (59,807) | ||
| SETDB1 | K-562 | ENCODE | Homo sapiens | ENCSR000AUT | Download (500) | ||
| SETDB1 | K-562 | ENCODE | Homo sapiens | ENCSR000EWD | Download (6,361) | ||
| SETDB1 | K-562 | ENCODE | Homo sapiens | ENCSR000EWI | Download (19,731) | ||
| SETDB1 | WN8532 | GEO | Homo sapiens | GSE36579 | Download (5,283) | ||
| SETX | A-549 | Influenza_PR8_NS1 | GEO | Homo sapiens | GSE52936 | Download (11,008) | |
| SFMBT1 | HeLa-S3 | G1 | GEO | Homo sapiens | GSE45441 | Download (11) | |
| SFMBT1 | HeLa | GEO | Homo sapiens | GSE45441 | Download (8,675) | ||
| SFPQ | LTAD | DHT-1nM | GEO | Homo sapiens | GSE94577 | Download (5,297) | |
| SFPQ | LTAD | EtOH | GEO | Homo sapiens | GSE94577 | Download (9,243) | |
| SFPQ | LTAD | siCTBP1-AS-DH | GEO | Homo sapiens | GSE94577 | Download (145) | |
| SFPQ | LTAD | siCTBP1-AS-EtOH | GEO | Homo sapiens | GSE94577 | Download (6,724) | |
| SGF29 | HeLa | GEO | Homo sapiens | GSE20303 | Download (141) | ||
| SGR5 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (482) |
| SHL | Col-0 | Col-0_seedling | 10d | GEO | Arabidopsis thaliana | GSE104369 | Download (2,539) |
| SIN3A | GM12878 | ENCODE | Homo sapiens | ENCSR000DYX | Download (2,423) | ||
| SIN3A | PFSK1 | ENCODE | Homo sapiens | ENCSR000BOY | Download (13,121) | ||
| SIN3A | A-549 | ENCODE | Homo sapiens | ENCSR000BRM | Download (31,168) | ||
| SIN3A | A-549 | ENCODE | Homo sapiens | ENCSR513XQX | Download (33,422) | ||
| SIN3A | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BGL | Download (35,893) | ||
| SIN3A | HUVEC-C | GEO | Homo sapiens | GSE103245 | Download (40,538) | ||
| SIN3A | K-562 | ENCODE | Homo sapiens | ENCSR000BLR | Download (14,138) | ||
| SIN3A | K-562 | ENCODE | Homo sapiens | ENCSR920BLG | Download (24,617) | ||
| SIN3A | MCF-7 | ENCODE | Homo sapiens | ENCSR000BUM | Download (49,667) | ||
| SIN3A | MCF-7 | ENCODE | Homo sapiens | ENCSR468LUO | Download (32,386) | ||
| SIN3A | PANC-1 | ENCODE | Homo sapiens | ENCSR000BOW | Download (25,965) | ||
| SIN3A | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BPB | Download (71,035) | ||
| SIN3A | WA01 | ENCODE | Homo sapiens | ENCSR000BIS | Download (32,054) | ||
| SIN3A | WA01 | ENCODE | Homo sapiens | ENCSR000EBO | Download (47,400) | ||
| SIN3A | HCT-116 | ENCODE | Homo sapiens | ENCSR000BSG | Download (11,509) | ||
| SIN3B | K-562 | ENCODE | Homo sapiens | ENCSR657JLK | Download (5,037) | ||
| SIN3B | K-562 | ENCODE | Homo sapiens | ENCSR887ZEN | Download (9,701) | ||
| SIN3B | Hep-G2 | ENCODE | Homo sapiens | ENCSR452YHM | Download (23,073) | ||
| SIRT3 | U2OS | GEO | Homo sapiens | GSE41000 | Download (55) | ||
| SIRT3 | U2OS | KOSIRT3 | GEO | Homo sapiens | GSE41000 | Download (1) | |
| SIRT3 | U2OS | UV | GEO | Homo sapiens | GSE41000 | Download (2) | |
| SIRT6 | K-562 | ENCODE | Homo sapiens | ENCSR000AUB | Download (9,165) | ||
| SIRT6 | K-562 | ENCODE | Homo sapiens | ENCSR000DOH | Download (2,220) | ||
| SIRT6 | SK-MEL-239 | L-C-B | GEO | Homo sapiens | GSE102813 | Download (51,377) | |
| SIRT6 | SK-MEL-239 | SIRT6-2-7 | GEO | Homo sapiens | GSE102813 | Download (3,797) | |
| SIRT6 | WA01 | ENCODE | Homo sapiens | ENCSR000AUS | Download (1,538) | ||
| SIX2 | HEK | GEO | Homo sapiens | GSE73865 | Download (57,534) | ||
| SIX2 | kidney | fetal_16w | GEO | Homo sapiens | GSE75948 | Download (25,980) | |
| SIX2 | kidney | fetal_17w | GEO | Homo sapiens | GSE75948 | Download (52,424) | |
| SIX4 | MCF-7 | ENCODE | Homo sapiens | ENCSR279IEM | Download (445) | ||
| SIX5 | WA01 | ENCODE | Homo sapiens | ENCSR000BIQ | Download (3,044) | ||
| SIX5 | GM12878 | ENCODE | Homo sapiens | ENCSR000BJE | Download (10,626) | ||
| SIX5 | A-549 | ENCODE | Homo sapiens | ENCSR000BRL | Download (17,669) | ||
| SIX5 | K-562 | ENCODE | Homo sapiens | ENCSR000BGX | Download (2,231) | ||
| SIX5 | K-562 | ENCODE | Homo sapiens | ENCSR000BNW | Download (3,035) | ||
| SKI | Hep-G2 | ENCODE | Homo sapiens | ENCSR754MUD | Download (38,811) | ||
| SKI | HL-60 | GEO | Homo sapiens | GSE107553 | Download (43,968) | ||
| SKI | HL-60 | CRISPRCas9_ctrl | GEO | Homo sapiens | GSE107553 | Download (21,940) | |
| SKIL | GM12878 | ENCODE | Homo sapiens | ENCSR212YKD | Download (48,361) | ||
| SKIL | K-562 | ENCODE | Homo sapiens | ENCSR336DXE | Download (19,205) | ||
| SMAD1 | GM12878 | ENCODE | Homo sapiens | ENCSR813DCK | Download (15,788) | ||
| SMAD1 | K-562 | ENCODE | Homo sapiens | ENCSR038DJJ | Download (25,016) | ||
| SMAD1 | BGO3 | GEO | Homo sapiens | GSE36578 | Download (26,990) | ||
| SMAD1 | CD34 | ERYTH_BMP | GEO | Homo sapiens | GSE29194 | Download (3,670) | |
| SMAD1 | CD34 | PROG_BMP | GEO | Homo sapiens | GSE29194 | Download (24,506) | |
| SMAD1-5 | MDA-MB-231 | TGF-beta | GEO | Homo sapiens | GSE92443 | Download (1,862) | |
| SMAD1-5-8 | A-375 | GEO | Homo sapiens | GSE83347 | Download (568) | ||
| SMAD2 | HASMC | PBS | GEO | Homo sapiens | GSE112326 | Download (472) | |
| SMAD2 | HASMC | TGFb | GEO | Homo sapiens | GSE112326 | Download (1,075) | |
| SMAD2 | hESC | activinA_15h | GEO | Homo sapiens | GSE99202 | Download (4,380) | |
| SMAD2 | hESC | YAP-_activinA_15h | GEO | Homo sapiens | GSE99202 | Download (7,296) | |
| SMAD2 | endoderm | GEO | Homo sapiens | GSE29422 | Download (7,541) | ||
| SMAD2 | hESC | GEO | Homo sapiens | GSE29422 | Download (2,943) | ||
| SMAD2 | WA09 | DMSO | GEO | Homo sapiens | GSE52437 | Download (8) | |
| SMAD2 | K-562 | ENCODE | Homo sapiens | ENCSR189PYJ | Download (392) | ||
| SMAD2-3 | MCF-10A | TGFB-16h | GEO | Homo sapiens | GSE83787 | Download (785) | |
| SMAD3 | HCASMC | GEO | Homo sapiens | GSE115317 | Download (188) | ||
| SMAD3 | NCI-H441 | GEO | Homo sapiens | GSE51509 | Download (49,883) | ||
| SMAD3 | NCI-H441 | SIITTF1 | GEO | Homo sapiens | GSE51509 | Download (54,826) | |
| SMAD3 | HCC1954 | GEO | Homo sapiens | GSE104760 | Download (61,772) | ||
| SMAD3 | HCC1954 | TGFb | GEO | Homo sapiens | GSE104760 | Download (65,263) | |
| SMAD3 | MDA-MB-231 | GEO | Homo sapiens | GSE104760 | Download (106,365) | ||
| SMAD3 | MDA-MB-231 | TGFb | GEO | Homo sapiens | GSE104760 | Download (110,217) | |
| SMAD3 | HMLE | GEO | Homo sapiens | GSE104760 | Download (879) | ||
| SMAD3 | HMLE | Doxycicline_TGFb | GEO | Homo sapiens | GSE104760 | Download (14,940) | |
| SMAD3 | HMLE | TGFb | GEO | Homo sapiens | GSE104760 | Download (4,633) | |
| SMAD3 | BGO3 | GEO | Homo sapiens | GSE21614 | Download (17,097) | ||
| SMAD3 | BGO3 | GEO | Homo sapiens | GSE36578 | Download (34,287) | ||
| SMAD3 | BGO3 | DIFF_0H | GEO | Homo sapiens | GSE36578 | Download (2,615) | |
| SMAD3 | BGO3 | DIFF_2H | GEO | Homo sapiens | GSE36578 | Download (8,877) | |
| SMAD3 | BGO3 | DIFF_48H | GEO | Homo sapiens | GSE36578 | Download (8,318) | |
| SMAD3 | endoderm | GEO | Homo sapiens | GSE29422 | Download (4,526) | ||
| SMAD3 | hESC | GEO | Homo sapiens | GSE29422 | Download (7,031) | ||
| SMAD3 | hESC | GEO | Homo sapiens | GSE75297 | Download (160) | ||
| SMAD3 | hESC | DIFF_D0 | GEO | Homo sapiens | GSE75297 | Download (89) | |
| SMAD3 | hESC | DIFF_D1 | GEO | Homo sapiens | GSE75297 | Download (277) | |
| SMAD3 | hESC | DIFF_D2 | GEO | Homo sapiens | GSE75297 | Download (103) | |
| SMAD3 | LX2 | CALCIPOTRIOL_TGFB1 | GEO | Homo sapiens | GSE38103 | Download (29,626) | |
| SMAD3 | LX2 | TGFB1 | GEO | Homo sapiens | GSE38103 | Download (1,235) | |
| SMAD3 | MDA-MB-231 | GEO | Homo sapiens | GSE92443 | Download (170) | ||
| SMAD3 | MDA-MB-231 | TGF-beta | GEO | Homo sapiens | GSE92443 | Download (5,119) | |
| SMAD4 | hESC | GEO | Homo sapiens | GSE29422 | Download (3,413) | ||
| SMAD4 | Hep-G2 | ENCODE | Homo sapiens | ENCSR826YMT | Download (123,370) | ||
| SMAD4 | Hep-G2 | Ab_13-2-1A5 | GEO | Homo sapiens | GSE97661 | Download (1,978) | |
| SMAD4 | Hep-G2 | Ab_R516-1-1G12 | GEO | Homo sapiens | GSE97661 | Download (11,483) | |
| SMAD4 | Hep-G2 | Ab_R516 | GEO | Homo sapiens | GSE97661 | Download (4,776) | |
| SMAD4 | endoderm | GEO | Homo sapiens | GSE29422 | Download (7,575) | ||
| SMAD5 | GM12878 | ENCODE | Homo sapiens | ENCSR251OVJ | Download (15,450) | ||
| SMAD5 | K-562 | ENCODE | Homo sapiens | ENCSR000FCD | Download (34,601) | ||
| SMARCA2 | SK-N-MC | shEWSFLI1 | GEO | Homo sapiens | GSE94275 | Download (15,923) | |
| SMARCA2 | SK-N-MC | shGFP | GEO | Homo sapiens | GSE94275 | Download (3,570) | |
| SMARCA4 | BT-16 | Dox | GEO | Homo sapiens | GSE71504 | Download (18,444) | |
| SMARCA4 | BT-16 | NoDox | GEO | Homo sapiens | GSE71504 | Download (1,430) | |
| SMARCA4 | G-401 | Dox | GEO | Homo sapiens | GSE71504 | Download (918) | |
| SMARCA4 | G-401 | NoDox | GEO | Homo sapiens | GSE71504 | Download (509) | |
| SMARCA4 | MCF-10A | GEO | Homo sapiens | GSE74716 | Download (27,092) | ||
| SMARCA4 | TTC-549 | Dox | GEO | Homo sapiens | GSE71504 | Download (6,701) | |
| SMARCA4 | TTC-549 | NoDox | GEO | Homo sapiens | GSE71504 | Download (6,134) | |
| SMARCA4 | WA09 | GEO | Homo sapiens | GSE105028 | Download (1,792) | ||
| SMARCA4 | Aska-SS | GEO | Homo sapiens | GSE108025 | Download (35,450) | ||
| SMARCA4 | Aska-SS | shSSX | GEO | Homo sapiens | GSE108025 | Download (47,237) | |
| SMARCA4 | K-562 | ENCODE | Homo sapiens | ENCSR587OQL | Download (74,342) | ||
| SMARCA4 | K-562 | ENCODE | Homo sapiens | ENCSR643VTW | Download (61,980) | ||
| SMARCA4 | SYO-1 | GEO | Homo sapiens | GSE108025 | Download (16,971) | ||
| SMARCA4 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EZC | Download (5,175) | ||
| SMARCA4 | neuron | bipolar_doxy_4d | ENCODE | Homo sapiens | ENCSR839SFJ | Download (109,209) | |
| SMARCA4 | J-Lat | GFP-Clone-A72_DMSO | GEO | Homo sapiens | GSE100266 | Download (23,208) | |
| SMARCA4 | J-Lat | GFP-Clone-A72_JQ1 | GEO | Homo sapiens | GSE100266 | Download (9,373) | |
| SMARCA4 | 501MEL | SHCTR | GEO | Homo sapiens | GSE61965 | Download (50,412) | |
| SMARCA4 | 501MEL | SHMITF | GEO | Homo sapiens | GSE61965 | Download (25,635) | |
| SMARCA4 | 501MEL | SHSOX10 | GEO | Homo sapiens | GSE61965 | Download (40,267) | |
| SMARCA4 | HCT-116 | GEO | Homo sapiens | GSE71510 | Download (1,883) | ||
| SMARCA5 | GM12878 | ENCODE | Homo sapiens | ENCSR706YUH | Download (51,298) | ||
| SMARCA5 | K-562 | ENCODE | Homo sapiens | ENCSR895HSJ | Download (34,014) | ||
| SMARCA5 | MCF-7 | ENCODE | Homo sapiens | ENCSR487ASM | Download (13,589) | ||
| SMARCB1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EDK | Download (55,182) | ||
| SMARCB1 | Hep-G2 | GEO | Homo sapiens | GSE69566 | Download (25,179) | ||
| SMARCB1 | K-562 | ENCODE | Homo sapiens | ENCSR000EHN | Download (2,735) | ||
| SMARCC1 | Aska-SS | GEO | Homo sapiens | GSE108025 | Download (54,976) | ||
| SMARCC1 | Aska-SS | shSSX | GEO | Homo sapiens | GSE108025 | Download (53,016) | |
| SMARCC1 | Hep-G2 | GEO | Homo sapiens | GSE108514 | Download (25,868) | ||
| SMARCC1 | HS-SY-2 | GEO | Homo sapiens | GSE108025 | Download (51,135) | ||
| SMARCC1 | SYO-1 | GEO | Homo sapiens | GSE108025 | Download (7,829) | ||
| SMARCC1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EDM | Download (29,862) | ||
| SMARCC1 | HCT-116 | GEO | Homo sapiens | GSE71510 | Download (2,347) | ||
| SMARCC1 | BT-16 | Dox | GEO | Homo sapiens | GSE71504 | Download (4,414) | |
| SMARCC1 | G-401 | Dox | GEO | Homo sapiens | GSE71504 | Download (4,535) | |
| SMARCC1 | G-401 | NoDox | GEO | Homo sapiens | GSE71504 | Download (1,804) | |
| SMARCC1 | SK-N-MC | GEO | Homo sapiens | GSE94275 | Download (38,119) | ||
| SMARCC1 | SK-N-MC | shEWSFLI1 | GEO | Homo sapiens | GSE94275 | Download (18,141) | |
| SMARCC1 | SK-N-MC | shGFP | GEO | Homo sapiens | GSE94275 | Download (17,358) | |
| SMARCC1 | TTC-549 | Dox | GEO | Homo sapiens | GSE71504 | Download (24,952) | |
| SMARCC1 | TTC-549 | NoDox | GEO | Homo sapiens | GSE71504 | Download (20,600) | |
| SMARCC2 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EDL | Download (5,945) | ||
| SMARCC2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR887LYD | Download (3,057) | ||
| SMARCC2 | K-562 | ENCODE | Homo sapiens | ENCSR519WMW | Download (16,165) | ||
| SMARCE1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR968QDP | Download (608) | ||
| SMARCE1 | K-562 | ENCODE | Homo sapiens | ENCSR157TCS | Download (63,058) | ||
| SMARCE1 | MCF-7 | ENCODE | Homo sapiens | ENCSR431TLD | Download (30,084) | ||
| SMARCE1 | HMLE-Twist-ER | 125nM_4OHT | GEO | Homo sapiens | GSE96933 | Download (14,064) | |
| SMB | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (5,934) |
| SMC1 | HAP1 | GEO | Homo sapiens | GSE94992 | Download (18,395) | ||
| SMC1 | HAP1 | SCC4KO | GEO | Homo sapiens | GSE94992 | Download (5,853) | |
| SMC1 | HAP1 | WaplKO-33 | GEO | Homo sapiens | GSE94992 | Download (39,289) | |
| SMC1 | HAP1 | WaplKO-33_SCC4KO | GEO | Homo sapiens | GSE94992 | Download (28,258) | |
| SMC1 | HMEC-1 | GEO | Homo sapiens | GSE101921 | Download (24,948) | ||
| SMC1 | MCF-10A | GEO | Homo sapiens | GSE101921 | Download (26,185) | ||
| SMC1 | HCAEC | GEO | Homo sapiens | GSE101921 | Download (75,631) | ||
| SMC1 | HCT-116 | RAD21-mAC | GEO | Homo sapiens | GSE104888 | Download (41,374) | |
| SMC1 | HCT-116 | RAD21-mAC_500uM_auxin | GEO | Homo sapiens | GSE104888 | Download (7,079) | |
| SMC1A | HCT-116 | GEO | Homo sapiens | GSE112000 | Download (13,752) | ||
| SMC1A | primary-epidermal-keratinocyte | diff_d0 | GEO | Homo sapiens | GSE84657 | Download (34,702) | |
| SMC1A | primary-epidermal-keratinocyte | diff_d3 | GEO | Homo sapiens | GSE84657 | Download (43,349) | |
| SMC1A | primary-epidermal-keratinocyte | diff_d6 | GEO | Homo sapiens | GSE84657 | Download (46,339) | |
| SMC1A | Primary-epidermal-keratinocyte | GEO | Homo sapiens | GSE85526 | Download (54,423) | ||
| SMC1A | monocyte | GEO | Homo sapiens | GSE98367 | Download (98,345) | ||
| SMC1A | monocyte | INFg | GEO | Homo sapiens | GSE98367 | Download (71,927) | |
| SMC1A | LCL | GEO | Homo sapiens | GSE38395 | Download (18,547) | ||
| SMC1A | BCBL-1 | GEO | Homo sapiens | GSE38411 | Download (101,124) | ||
| SMC1A | A-549 | GEO | Homo sapiens | GSE76893 | Download (62,895) | ||
| SMC1A | Hep-G2 | GEO | Homo sapiens | GSE76893 | Download (65,889) | ||
| SMC1A | MCF-7 | GEO | Homo sapiens | GSE76893 | Download (43,086) | ||
| SMC1A | GM12878 | GEO | Homo sapiens | GSE93080 | Download (84,565) | ||
| SMC3 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000EHW | Download (36,183) | ||
| SMC3 | U-937 | ENA | Homo sapiens | ERP008568 | Download (77) | ||
| SMC3 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECS | Download (50,771) | ||
| SMC3 | HeLa-Kyoto | GEO | Homo sapiens | GSE102884 | Download (74,610) | ||
| SMC3 | HeLa-Kyoto | PDS5-depleted | GEO | Homo sapiens | GSE102884 | Download (67,468) | |
| SMC3 | HeLa-Kyoto | WAPL-depleted | GEO | Homo sapiens | GSE102884 | Download (86,935) | |
| SMC3 | HeLa-Kyoto | WAPL_PDS-depleted | GEO | Homo sapiens | GSE102884 | Download (69,231) | |
| SMC3 | GP5D | GEO | Homo sapiens | GSE51234 | Download (60,750) | ||
| SMC3 | GP5D | SIRAD21 | GEO | Homo sapiens | GSE51234 | Download (13,405) | |
| SMC3 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZP | Download (15,056) | ||
| SMC3 | HeLa | ESCO2_ctrl_siRNA | GEO | Homo sapiens | GSE105004 | Download (836) | |
| SMC3 | HeLa | ESCO2-deltaPBMA_ctrl_siRNA | GEO | Homo sapiens | GSE105004 | Download (236) | |
| SMC3 | HeLa | ESCO2-deltaPBMA_ctrl_siRNA | GEO | Homo sapiens | GSE105004 | Download (20,088) | |
| SMC3 | HeLa | ESCO2-deltaPBMA_ESCO1_siRNA | GEO | Homo sapiens | GSE105004 | Download (18,488) | |
| SMC3 | HeLa | ESCO2_ESCO1_siRNA | GEO | Homo sapiens | GSE105004 | Download (11,733) | |
| SMC3 | neural | ENCODE | Homo sapiens | ENCSR404BPV | Download (48,480) | ||
| SMC3 | A-549 | ENCODE | Homo sapiens | ENCSR481YWD | Download (28,281) | ||
| SMC3 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EDW | Download (37,842) | ||
| SMC3 | HL-60 | ENA | Homo sapiens | ERP008568 | Download (130,008) | ||
| SMC3 | IMR-90 | ENCODE | Homo sapiens | ENCSR000HPG | Download (59,776) | ||
| SMC3 | K-562 | ENCODE | Homo sapiens | ENCSR000EGW | Download (36,921) | ||
| SMC4 | HeLa | M | GEO | Homo sapiens | GSE22478 | Download (139) | |
| SNAI1 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (132) | ||
| SNAI2 | keratinocyte | LacZ | GEO | Homo sapiens | GSE55421 | Download (25,014) | |
| SNAI2 | keratinocyte | LacZ_DIFF | GEO | Homo sapiens | GSE55421 | Download (4,424) | |
| SNAI2 | keratinocyte | SHCTR | GEO | Homo sapiens | GSE55421 | Download (7,108) | |
| SNAI2 | keratinocyte | SHSNAI2 | GEO | Homo sapiens | GSE55421 | Download (879) | |
| SNAI2 | keratinocyte | SNAI2 | GEO | Homo sapiens | GSE55421 | Download (12,485) | |
| SNAI2 | keratinocyte | SNAI2_diff | GEO | Homo sapiens | GSE55421 | Download (80,558) | |
| SNAPC1 | MCF-10A | GEO | Homo sapiens | GSE37403 | Download (3,642) | ||
| SNAPC4 | MCF-10A | GEO | Homo sapiens | GSE37403 | Download (1,426) | ||
| SND1 | NHEK | GEO | Homo sapiens | GSE29498 | Download (14,944) | ||
| SNIP1 | K-562 | ENCODE | Homo sapiens | ENCSR654CQU | Download (2,000) | ||
| SNIP1 | MCF-7 | ENCODE | Homo sapiens | ENCSR042TWZ | Download (1,804) | ||
| SNRNP70 | K-562 | ENCODE | Homo sapiens | ENCSR754ZHU | Download (4,701) | ||
| SOC1 | Col-0 | Col-0_whole-plant | 18d | GEO | Arabidopsis thaliana | GSE45846 | Download (1,537) |
| SOX10 | 501MEL | GEO | Homo sapiens | GSE61965 | Download (4,198) | ||
| SOX11 | GRANT-A519 | GEO | Homo sapiens | GSE52146 | Download (38,769) | ||
| SOX13 | Hep-G2 | ENCODE | Homo sapiens | ENCSR445ACU | Download (17,188) | ||
| SOX13 | Hep-G2 | ENCODE | Homo sapiens | ENCSR715UCI | Download (116,256) | ||
| SOX2 | hiPSC | GEO | Homo sapiens | GSE56567 | Download (64,223) | ||
| SOX2 | hiPSC | GEO | Homo sapiens | GSE67282 | Download (6,645) | ||
| SOX2 | hiPSC | INHI | GEO | Homo sapiens | GSE67282 | Download (3,238) | |
| SOX2 | hiPSC | KDP53 | GEO | Homo sapiens | GSE67282 | Download (5,025) | |
| SOX2 | hiPSC | KDP53_INHI | GEO | Homo sapiens | GSE67282 | Download (6,198) | |
| SOX2 | hiPSC | 3s2 | GEO | Homo sapiens | GSE81899 | Download (879) | |
| SOX2 | OSK | GEO | Homo sapiens | GSE81899 | Download (1,237) | ||
| SOX2 | OSKM | GEO | Homo sapiens | GSE81899 | Download (5,507) | ||
| SOX2 | OSvK | GEO | Homo sapiens | GSE81899 | Download (263) | ||
| SOX2 | OSvKM | GEO | Homo sapiens | GSE81899 | Download (257) | ||
| SOX2 | BJ | INDUCED | GEO | Homo sapiens | GSE36570 | Download (121,462) | |
| SOX2 | glioma | stem | GEO | Homo sapiens | GSE67282 | Download (3,859) | |
| SOX2 | hESC | GEO | Homo sapiens | GSE18292 | Download (18,963) | ||
| SOX2 | hESC | GEO | Homo sapiens | GSE69479 | Download (2,352) | ||
| SOX2 | HNSC | GEO | Homo sapiens | GSE69479 | Download (74,636) | ||
| SOX2 | KYSE70 | GEO | Homo sapiens | GSE46837 | Download (6,840) | ||
| SOX2 | RENVM | GEO | Homo sapiens | GSE49404 | Download (38,149) | ||
| SOX2 | RENVM | SHSOX2 | GEO | Homo sapiens | GSE49404 | Download (4,497) | |
| SOX2 | TT | GEO | Homo sapiens | GSE46837 | Download (10,181) | ||
| SOX2 | H9 | GEO | Homo sapiens | GSE46837 | Download (3,216) | ||
| SOX4 | MRC-5 | 1DPT | GEO | Homo sapiens | GSE75910 | Download (19,682) | |
| SOX4 | HCC1954 | GEO | Homo sapiens | GSE104760 | Download (7,814) | ||
| SOX4 | HCC1954 | TGFb | GEO | Homo sapiens | GSE104760 | Download (3,565) | |
| SOX4 | MDA-MB-231 | GEO | Homo sapiens | GSE104760 | Download (19,732) | ||
| SOX4 | MDA-MB-231 | TGFb | GEO | Homo sapiens | GSE104760 | Download (15,057) | |
| SOX4 | HMLE | GEO | Homo sapiens | GSE104760 | Download (180) | ||
| SOX4 | HMLE | Doxycicline | GEO | Homo sapiens | GSE104760 | Download (251) | |
| SOX4 | HMLE | Doxycicline_TGFb | GEO | Homo sapiens | GSE104760 | Download (682) | |
| SOX4 | HMLE | TGFb | GEO | Homo sapiens | GSE104760 | Download (570) | |
| SOX5 | Hep-G2 | ENCODE | Homo sapiens | ENCSR961WLZ | Download (99,396) | ||
| SOX6 | Hep-G2 | ENCODE | Homo sapiens | ENCSR543BVU | Download (2,005) | ||
| SOX6 | K-562 | ENCODE | Homo sapiens | ENCSR788RSW | Download (46,163) | ||
| SOX9 | HT29 | GEO | Homo sapiens | GSE63629 | Download (20,238) | ||
| SP1 | A-375 | A771726 | GEO | Homo sapiens | GSE68044 | Download (3,750) | |
| SP1 | A-375 | DMSO | GEO | Homo sapiens | GSE68044 | Download (10,313) | |
| SP1 | K-562 | ENCODE | Homo sapiens | ENCSR991ELG | Download (8,785) | ||
| SP1 | MCF-7 | ENCODE | Homo sapiens | ENCSR729LGA | Download (2,605) | ||
| SP1 | WA01 | ENCODE | Homo sapiens | ENCSR000BIR | Download (37,928) | ||
| SP1 | HCT-116 | ENCODE | Homo sapiens | ENCSR000BSF | Download (28,155) | ||
| SP1 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (28,769) | ||
| SP1 | GM12878 | ENCODE | Homo sapiens | ENCSR000BHK | Download (44,580) | ||
| SP1 | HEK293T | ENCODE | Homo sapiens | ENCSR906PEI | Download (350) | ||
| SP1 | HEK293T | ENA | Homo sapiens | ERP007114 | Download (14,645) | ||
| SP1 | liver | ENCODE | Homo sapiens | ENCSR085IXF | Download (105,143) | ||
| SP1 | liver | ENCODE | Homo sapiens | ENCSR386YIH | Download (83,895) | ||
| SP1 | A-549 | ENCODE | Homo sapiens | ENCSR000BPE | Download (51,962) | ||
| SP1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BJX | Download (30,372) | ||
| SP1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR334KIQ | Download (1,280) | ||
| SP1 | HL-60 | ENA | Homo sapiens | ERP008568 | Download (200) | ||
| SP1 | K-562 | ENCODE | Homo sapiens | ENCSR000BKO | Download (3,160) | ||
| SP140 | macrophage | GEO | Homo sapiens | GSE90126 | Download (154,461) | ||
| SP140 | macrophage | LPS | GEO | Homo sapiens | GSE90126 | Download (247,323) | |
| SP2 | HEK293 | ENCODE | Homo sapiens | ENCSR807LQP | Download (31,764) | ||
| SP2 | HEK293T | ENA | Homo sapiens | ERP007114 | Download (10,808) | ||
| SP2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BOU | Download (1,034) | ||
| SP2 | K-562 | ENCODE | Homo sapiens | ENCSR000BNL | Download (5,297) | ||
| SP2 | WA01 | ENCODE | Homo sapiens | ENCSR000BQG | Download (2,252) | ||
| SP2 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (34,893) | ||
| SP3 | HEK293 | ENCODE | Homo sapiens | ENCSR141PZA | Download (25,531) | ||
| SP4 | WA01 | ENCODE | Homo sapiens | ENCSR000BQV | Download (26,809) | ||
| SP4 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (37,589) | ||
| SP5 | Hep-G2 | ENCODE | Homo sapiens | ENCSR019NPF | Download (54,487) | ||
| SP7 | HEK293 | ENCODE | Homo sapiens | ENCSR468IJT | Download (70,423) | ||
| SPCH | Col | Col_seedling | 4d | GEO | Arabidopsis thaliana | GSE57497 | Download (21,502) |
| SPDEF | A-549 | GEO | Homo sapiens | GSE86957 | Download (11,577) | ||
| SPI1 | macrophage | IFNG | GEO | Homo sapiens | GSE47188 | Download (12,221) | |
| SPI1 | macrophage | IL4 | GEO | Homo sapiens | GSE47188 | Download (4,934) | |
| SPI1 | macrophage | IFNG | GEO | Homo sapiens | GSE66594 | Download (170,070) | |
| SPI1 | macrophage | IL4 | GEO | Homo sapiens | GSE66594 | Download (188,903) | |
| SPI1 | macrophage | TPP | GEO | Homo sapiens | GSE66594 | Download (125,888) | |
| SPI1 | monocyte | GEO | Homo sapiens | GSE31621 | Download (53,628) | ||
| SPI1 | monocyte | MACROPHAGE | GEO | Homo sapiens | GSE31621 | Download (43,671) | |
| SPI1 | K-562 | GEO | Homo sapiens | GSE70482 | Download (62,723) | ||
| SPI1 | K-562 | GEO | Homo sapiens | GSE74999 | Download (2,161) | ||
| SPI1 | K-562 | NABUT | GEO | Homo sapiens | GSE74999 | Download (40,350) | |
| SPI1 | K-562 | SAHA | GEO | Homo sapiens | GSE74999 | Download (10,865) | |
| SPI1 | Kasumi-1 | SICTR | GEO | Homo sapiens | GSE60130 | Download (54,806) | |
| SPI1 | Kasumi-1 | SIRUNX1ETO | GEO | Homo sapiens | GSE60130 | Download (51,842) | |
| SPI1 | NCI-H929 | GEO | Homo sapiens | GSE56857 | Download (30,333) | ||
| SPI1 | OCI-Ly10 | GEO | Homo sapiens | GSE56857 | Download (7,214) | ||
| SPI1 | OCI-Ly3 | GEO | Homo sapiens | GSE56857 | Download (3,596) | ||
| SPI1 | OCI-Ly3 | SHCTR | GEO | Homo sapiens | GSE56857 | Download (3,030) | |
| SPI1 | OCI-Ly3 | SHSPIB | GEO | Homo sapiens | GSE56857 | Download (641) | |
| SPI1 | OCI-Ly7 | GEO | Homo sapiens | GSE69558 | Download (36,121) | ||
| SPI1 | THP-1 | DIFF | GEO | Homo sapiens | GSE25426 | Download (27,256) | |
| SPI1 | THP-1 | 1-25-OH-2D3 | GEO | Homo sapiens | GSE89178 | Download (229,174) | |
| SPI1 | THP-1 | EtOH | GEO | Homo sapiens | GSE89178 | Download (212,938) | |
| SPI1 | AML | GEO | Homo sapiens | GSE112074 | Download (131,555) | ||
| SPI1 | AML | OG86 | GEO | Homo sapiens | GSE112074 | Download (122,651) | |
| SPI1 | ME-1 | GEO | Homo sapiens | GSE46044 | Download (16,909) | ||
| SPI1 | RS4-11 | GEO | Homo sapiens | GSE71616 | Download (10,201) | ||
| SPI1 | RS4-11 | DEX | GEO | Homo sapiens | GSE71616 | Download (17,789) | |
| SPI1 | monocyte | GEO | Homo sapiens | GSE98367 | Download (140,946) | ||
| SPI1 | monocyte | INFg | GEO | Homo sapiens | GSE98367 | Download (200,720) | |
| SPI1 | GM12878 | ENCODE | Homo sapiens | ENCSR000BGQ | Download (80,881) | ||
| SPI1 | GM12891 | ENCODE | Homo sapiens | ENCSR000BIJ | Download (55,727) | ||
| SPI1 | LCL | ENA | Homo sapiens | ERP004110 | Download (183,775) | ||
| SPI1 | HL-60 | ENCODE | Homo sapiens | ENCSR000BUW | Download (92,220) | ||
| SPI1 | K-562 | ENCODE | Homo sapiens | ENCSR000BGW | Download (59,259) | ||
| SPI1 | THP-1 | eGFP-Pam3csk-0h | GEO | Homo sapiens | GSE103477 | Download (107,254) | |
| SPI1 | THP-1 | eGFP-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (110,932) | |
| SPI1 | THP-1 | NS1-Pam3csk-0h | GEO | Homo sapiens | GSE103477 | Download (93,734) | |
| SPI1 | THP-1 | NS1-Pam3csk-4h | GEO | Homo sapiens | GSE103477 | Download (89,857) | |
| SPI1 | U-937 | ENA | Homo sapiens | ERP008568 | Download (104,872) | ||
| SPI1 | MDM | dNS1 | GEO | Homo sapiens | GSE103477 | Download (141,944) | |
| SPI1 | MDM | H5N1 | GEO | Homo sapiens | GSE103477 | Download (143,742) | |
| SPI1 | MDM | IFNb | GEO | Homo sapiens | GSE103477 | Download (121,793) | |
| SPI1 | MDM | Mock | GEO | Homo sapiens | GSE103477 | Download (124,379) | |
| SPI1 | CD34 | ADULT | GEO | Homo sapiens | GSE70660 | Download (173,233) | |
| SPI1 | CD34 | FETAL | GEO | Homo sapiens | GSE70660 | Download (65,021) | |
| SPI1 | dendrite | GEO | Homo sapiens | GSE58864 | Download (14,655) | ||
| SPI1 | DOHH2 | GEO | Homo sapiens | GSE69558 | Download (83,343) | ||
| SPIB | OCI-Ly10 | GEO | Homo sapiens | GSE56857 | Download (18,095) | ||
| SPIB | OCI-Ly3 | GEO | Homo sapiens | GSE56857 | Download (28,001) | ||
| SPIB | OCI-Ly3 | SHCTR | GEO | Homo sapiens | GSE56857 | Download (28,168) | |
| SPIB | OCI-Ly3 | SHSPIB | GEO | Homo sapiens | GSE56857 | Download (3,497) | |
| SPIN1 | T778 | GEO | Homo sapiens | GSE57499 | Download (2,976) | ||
| SPL1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,007) |
| SPL1 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (6,111) |
| SPL13A | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (280) |
| SPL14 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,391) |
| SPL3 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (602) |
| SPL9 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,716) |
| SRC | MDA-MB-231 | GEO | Homo sapiens | GSE95121 | Download (288) | ||
| SRC | MDA-MB-231 | 45min | GEO | Homo sapiens | GSE95121 | Download (2,493) | |
| SRC | MDA-MB-231 | LQ | GEO | Homo sapiens | GSE95121 | Download (2,778) | |
| SRC | MDA-MB-231 | LQ_45min | GEO | Homo sapiens | GSE95121 | Download (3,590) | |
| SREBF1 | K-562 | ENCODE | Homo sapiens | ENCSR815ZDS | Download (12,832) | ||
| SREBF1 | MCF-7 | ENCODE | Homo sapiens | ENCSR197DJH | Download (5,574) | ||
| SREBF1 | GM12878 | ENCODE | Homo sapiens | ENCSR000DYU | Download (1,516) | ||
| SREBF1 | A-549 | ENCODE | Homo sapiens | ENCSR897MYK | Download (4,004) | ||
| SREBF1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEO | Download (1,467) | ||
| SREBF1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EZP | Download (273) | ||
| SREBF2 | GM12878 | ENCODE | Homo sapiens | ENCSR000DYT | Download (656) | ||
| SREBF2 | A-549 | ENCODE | Homo sapiens | ENCSR638QYO | Download (417) | ||
| SREBF2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EZO | Download (275) | ||
| SREBF2 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR611WZO | Download (22,972) | ||
| SRF | MCF-7 | ENCODE | Homo sapiens | ENCSR000BVA | Download (10,242) | ||
| SRF | WA01 | ENCODE | Homo sapiens | ENCSR000BIV | Download (5,916) | ||
| SRF | HCT-116 | ENCODE | Homo sapiens | ENCSR000BSC | Download (7,372) | ||
| SRF | A-673-clone-Asp114 | GEO | Homo sapiens | GSE92738 | Download (86,122) | ||
| SRF | GM12878 | ENCODE | Homo sapiens | ENCSR000BGE | Download (3,765) | ||
| SRF | GM12878 | ENCODE | Homo sapiens | ENCSR000BMI | Download (15,446) | ||
| SRF | GM12878 | ENCODE | Homo sapiens | ENCSR041XML | Download (14,594) | ||
| SRF | Ishikawa | ENCODE | Homo sapiens | ENCSR000BTD | Download (21,154) | ||
| SRF | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BLV | Download (13,722) | ||
| SRF | K-562 | ENCODE | Homo sapiens | ENCSR000BLK | Download (10,302) | ||
| SRM1 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (4,402) |
| SRSF3 | K-562 | ENCODE | Homo sapiens | ENCSR268QIQ | Download (22,314) | ||
| SRSF4 | Hep-G2 | ENCODE | Homo sapiens | ENCSR696MBC | Download (7,200) | ||
| SRSF7 | K-562 | ENCODE | Homo sapiens | ENCSR222MYK | Download (160) | ||
| SRSF9 | Hep-G2 | ENCODE | Homo sapiens | ENCSR624GNZ | Download (7,756) | ||
| SS18 | Aska-SS | GEO | Homo sapiens | GSE108025 | Download (38,236) | ||
| SS18 | Aska-SS | BAF47KO1 | GEO | Homo sapiens | GSE108025 | Download (30,156) | |
| SS18 | SYO-1 | GEO | Homo sapiens | GSE108025 | Download (36,011) | ||
| SS18 | SYO-1 | shSSX | GEO | Homo sapiens | GSE108025 | Download (19,407) | |
| SSRP1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR571PDN | Download (23,595) | ||
| SSRP1 | HT-1080 | AclacinomycinA | GEO | Homo sapiens | GSE107595 | Download (5,997) | |
| SSRP1 | HT-1080 | CBL0100 | GEO | Homo sapiens | GSE107595 | Download (5,673) | |
| SSRP1 | hiF-T | GEO | Homo sapiens | GSE98758 | Download (5,832) | ||
| STAG1 | MCF-10A | GEO | Homo sapiens | GSE101921 | Download (16,011) | ||
| STAG1 | MCF-10A | Control | GEO | Homo sapiens | GSE101921 | Download (4,104) | |
| STAG1 | MCF-10A | siSTAG2 | GEO | Homo sapiens | GSE101921 | Download (8,256) | |
| STAG1 | MCF-7 | ENA | Homo sapiens | ERP000209 | Download (73,519) | ||
| STAG1 | MCF-7 | E2 | ENA | Homo sapiens | ERP000209 | Download (77,113) | |
| STAG1 | MCF-7 | E2_SHCTCF | ENA | Homo sapiens | ERP000209 | Download (75,831) | |
| STAG1 | MCF-7 | E2_SHCTR | ENA | Homo sapiens | ERP000209 | Download (85,239) | |
| STAG1 | U-937 | ENA | Homo sapiens | ERP008568 | Download (11,130) | ||
| STAG1 | HCAEC | GEO | Homo sapiens | GSE101921 | Download (41,110) | ||
| STAG1 | erythroid | GEO | Homo sapiens | GSE67783 | Download (40,860) | ||
| STAG1 | HSPC | GEO | Homo sapiens | GSE67783 | Download (303,910) | ||
| STAG1 | CHRF28811 | ENA | Homo sapiens | ERP008568 | Download (34,291) | ||
| STAG1 | Hep-G2 | ENA | Homo sapiens | ERP000209 | Download (56,036) | ||
| STAG1 | HL-60 | ENA | Homo sapiens | ERP008568 | Download (23,185) | ||
| STAG1 | HMEC-1 | GEO | Homo sapiens | GSE101921 | Download (24,465) | ||
| STAG2 | HMEC-1 | GEO | Homo sapiens | GSE101921 | Download (31,934) | ||
| STAG2 | MCF-10A | GEO | Homo sapiens | GSE101921 | Download (21,808) | ||
| STAG2 | MCF-10A | Control | GEO | Homo sapiens | GSE101921 | Download (15,478) | |
| STAG2 | MCF-10A | siSTAG1 | GEO | Homo sapiens | GSE101921 | Download (27,374) | |
| STAG2 | MCF-10A | siSTAG2 | GEO | Homo sapiens | GSE101921 | Download (1,582) | |
| STAG2 | HCAEC | GEO | Homo sapiens | GSE101921 | Download (46,097) | ||
| STAT1 | SET-2 | GEO | Homo sapiens | GSE100566 | Download (77,817) | ||
| STAT1 | SET-2 | cortistatin-A | GEO | Homo sapiens | GSE100566 | Download (13,398) | |
| STAT1 | SET-2 | DMSO | GEO | Homo sapiens | GSE100566 | Download (25,458) | |
| STAT1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EZK | Download (18,141) | ||
| STAT1 | AGS | GEO | Homo sapiens | GSE79707 | Download (1,450) | ||
| STAT1 | CD14 | GEO | Homo sapiens | GSE43036 | Download (10,636) | ||
| STAT1 | CD14 | INFG | GEO | Homo sapiens | GSE43036 | Download (57,679) | |
| STAT1 | CD14 | INFG_LPS | GEO | Homo sapiens | GSE43036 | Download (67,375) | |
| STAT1 | CD14 | LPS | GEO | Homo sapiens | GSE43036 | Download (49,126) | |
| STAT1 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZM | Download (1) | ||
| STAT1 | GM12878 | ENCODE | Homo sapiens | ENCSR332EYT | Download (5,377) | ||
| STAT1 | K-562 | ENCODE | Homo sapiens | ENCSR000EHJ | Download (2,435) | ||
| STAT1 | K-562 | ENCODE | Homo sapiens | ENCSR000EHK | Download (1,266) | ||
| STAT1 | K-562 | ENCODE | Homo sapiens | ENCSR000FAU | Download (871) | ||
| STAT1 | K-562 | ENCODE | Homo sapiens | ENCSR000FAV | Download (592) | ||
| STAT1 | NCI-H358 | GEO | Homo sapiens | GSE79707 | Download (15,540) | ||
| STAT2 | GM12878 | GEO | Homo sapiens | GSE97661 | Download (3,412) | ||
| STAT2 | K-562 | ENCODE | Homo sapiens | ENCSR000FAT | Download (2,556) | ||
| STAT2 | K-562 | ENCODE | Homo sapiens | ENCSR000FBC | Download (1,872) | ||
| STAT3 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZV | Download (58) | ||
| STAT3 | U2932 | GEO | Homo sapiens | GSE50723 | Download (707) | ||
| STAT3 | HCC1143 | EtOH | GEO | Homo sapiens | GSE85579 | Download (17,661) | |
| STAT3 | HepaRG | GEO | Homo sapiens | GSE89157 | Download (141,053) | ||
| STAT3 | HepaRG | sodium-oleate | GEO | Homo sapiens | GSE89157 | Download (6,692) | |
| STAT3 | MCF10A-Er-Src | EtOH | GEO | Homo sapiens | GSE115597 | Download (33,316) | |
| STAT3 | MCF10A-Er-Src | TAM | GEO | Homo sapiens | GSE115597 | Download (66,181) | |
| STAT3 | MDA-MB-157 | EtOH | GEO | Homo sapiens | GSE85579 | Download (45,005) | |
| STAT3 | MDA-MB-231 | EtOH | GEO | Homo sapiens | GSE85579 | Download (35,162) | |
| STAT3 | MDA-MB-468 | EtOH | GEO | Homo sapiens | GSE85579 | Download (38,738) | |
| STAT3 | NCI-H358 | GEO | Homo sapiens | GSE79707 | Download (35,079) | ||
| STAT3 | OCI-Ly10 | GEO | Homo sapiens | GSE50723 | Download (74) | ||
| STAT3 | OCI-Ly19 | GEO | Homo sapiens | GSE50723 | Download (2,679) | ||
| STAT3 | OCI-Ly3 | GEO | Homo sapiens | GSE50723 | Download (8,890) | ||
| STAT3 | OCI-Ly7 | GEO | Homo sapiens | GSE50723 | Download (4,592) | ||
| STAT3 | SU-DHL-10 | GEO | Homo sapiens | GSE50723 | Download (2,563) | ||
| STAT3 | SU-DHL-2 | GEO | Homo sapiens | GSE50723 | Download (21,419) | ||
| STAT3 | SU-DHL-4 | GEO | Homo sapiens | GSE50723 | Download (3,172) | ||
| STAT3 | SU-DHL-6 | GEO | Homo sapiens | GSE50723 | Download (206) | ||
| STAT3 | MCF-10A | ENCODE | Homo sapiens | ENCSR000DOQ | Download (63,066) | ||
| STAT3 | MCF-10A | ENCODE | Homo sapiens | ENCSR000DOZ | Download (32,003) | ||
| STAT3 | MCF-10A | ENCODE | Homo sapiens | ENCSR000DPB | Download (86,444) | ||
| STAT3 | OCI-Ly10 | GEO | Homo sapiens | GSE106844 | Download (3,450) | ||
| STAT3 | OCI-Ly10 | AZD1480 | GEO | Homo sapiens | GSE106844 | Download (996) | |
| STAT3 | TMD8 | GEO | Homo sapiens | GSE106844 | Download (7,217) | ||
| STAT3 | TMD8 | AZD1480 | GEO | Homo sapiens | GSE106844 | Download (1,815) | |
| STAT3 | WA01 | ENA | Homo sapiens | ERP004237 | Download (59,351) | ||
| STAT3 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EDC | Download (20,309) | ||
| STAT3 | HCC70 | EtOH | GEO | Homo sapiens | GSE85579 | Download (76,929) | |
| STAT3 | Th17 | GEO | Homo sapiens | GSE67183 | Download (507) | ||
| STAT3 | A-137 | GEO | Homo sapiens | GSE85579 | Download (16,537) | ||
| STAT3 | A139 | GEO | Homo sapiens | GSE85579 | Download (16,963) | ||
| STAT5A | MV4-11 | GEO | Homo sapiens | GSE64862 | Download (20,817) | ||
| STAT5A | GM12878 | ENCODE | Homo sapiens | ENCSR000BQZ | Download (14,732) | ||
| STAT5A | K-562 | ENCODE | Homo sapiens | ENCSR000BRR | Download (16,454) | ||
| STAT5B | CD8 | GEO | Homo sapiens | GSE64713 | Download (3,197) | ||
| STAT5B | CD8 | H9 | GEO | Homo sapiens | GSE64713 | Download (28,174) | |
| STAT5B | CD8 | H9RET | GEO | Homo sapiens | GSE64713 | Download (10,091) | |
| STAT5B | CD8 | H9RETR | GEO | Homo sapiens | GSE64713 | Download (8,622) | |
| STAT5B | CD8 | IL2 | GEO | Homo sapiens | GSE64713 | Download (29,206) | |
| STKL2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (595) |
| STOP1 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (713) |
| SUPT16H | hiF-T | GEO | Homo sapiens | GSE98758 | Download (23,458) | ||
| SUPT20H | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECQ | Download (19) | ||
| SUPT20H | GM12878 | ENCODE | Homo sapiens | ENCSR000DNP | Download (1) | ||
| SUPT5H | HeLa | Flavo-0-H2O2 | GEO | Homo sapiens | GSE100742 | Download (3,826) | |
| SUPT5H | HeLa | Flavo-10min-H2O2 | GEO | Homo sapiens | GSE100742 | Download (2,715) | |
| SUPT5H | HeLa | Flavo-PJ34-0-H2O2 | GEO | Homo sapiens | GSE100742 | Download (6,422) | |
| SUPT5H | HeLa | Flavo-PJ34-10min-H2O2 | GEO | Homo sapiens | GSE100742 | Download (4,770) | |
| SUPT5H | MOLT-4 | GEO | Homo sapiens | GSE89384 | Download (95,388) | ||
| SUPT5H | MOLT-4 | NVP2 | GEO | Homo sapiens | GSE89384 | Download (23,150) | |
| SUPT5H | MOLT-4 | SNS | GEO | Homo sapiens | GSE89384 | Download (31,615) | |
| SUPT5H | K-562 | ENCODE | Homo sapiens | ENCSR894CGX | Download (103,429) | ||
| SUZ12 | MCF-7 | ENCODE | Homo sapiens | ENCSR757EMK | Download (11,801) | ||
| SUZ12 | NT2-D1 | ENCODE | Homo sapiens | ENCSR000EXH | Download (8,348) | ||
| SUZ12 | NT2-D1 | GEO | Homo sapiens | GSE101538 | Download (5,669) | ||
| SUZ12 | WA01 | ENCODE | Homo sapiens | ENCSR000ATS | Download (14,307) | ||
| SUZ12 | WA01 | ENCODE | Homo sapiens | ENCSR000EUQ | Download (5,934) | ||
| SUZ12 | CRL-7250 | shSS18 | GEO | Homo sapiens | GSE108025 | Download (27,025) | |
| SUZ12 | CRL-7250 | shSS18-SSX | GEO | Homo sapiens | GSE108025 | Download (17,251) | |
| SUZ12 | GM12878 | ENCODE | Homo sapiens | ENCSR091BOQ | Download (6,929) | ||
| SUZ12 | GM12878 | ENCODE | Homo sapiens | ENCSR744XTG | Download (8,350) | ||
| SUZ12 | HEK293T | ENCODE | Homo sapiens | ENCSR090ZDO | Download (236) | ||
| SUZ12 | Aska-SS | GEO | Homo sapiens | GSE108025 | Download (19,546) | ||
| SUZ12 | Hep-G2 | ENCODE | Homo sapiens | ENCSR771GTF | Download (3,480) | ||
| SUZ12 | K-562 | ENCODE | Homo sapiens | ENCSR000AUC | Download (16,970) | ||
| SUZ12 | K-562 | ENCODE | Homo sapiens | ENCSR412CTM | Download (13,855) | ||
| SUZ12 | Lu-130 | GEO | Homo sapiens | GSE99312 | Download (554) | ||
| SUZ12 | LNCaP | GEO | Homo sapiens | GSE39459 | Download (15,726) | ||
| SUZ12 | NCCIT | GEO | Homo sapiens | GSE71675 | Download (4,183) | ||
| SUZ12 | ProEs | GEO | Homo sapiens | GSE59087 | Download (23,499) | ||
| SUZ12 | LNCaP-abl | GEO | Homo sapiens | GSE39459 | Download (12,217) | ||
| SVIL | SH-SY5Y | B3_SHCTR | GEO | Homo sapiens | GSE58258 | Download (127) | |
| SVIL | SH-SY5Y | B3_SHSVIL | GEO | Homo sapiens | GSE58258 | Download (10) | |
| SVP | Col-0 | Col-0_seedling | 4d | GEO | Arabidopsis thaliana | GSE33120 | Download (14,734) |
| SVP | Col-0 | Col-0_seedling | 2w-FLC | GEO | Arabidopsis thaliana | GSE54881 | Download (8,337) |
| SVP | Col-0 | Col-0_seedling | 2w-flc-3 | GEO | Arabidopsis thaliana | GSE54881 | Download (2,312) |
| SVP | Col-0 | Col-0_inflorescence-flower-meristem | 2w-GFP | GEO | Arabidopsis thaliana | GSE33120 | Download (128) |
| SVP | Col-0 | Col-0_inflorescence-flower-meristem | 2w-wt | GEO | Arabidopsis thaliana | GSE33120 | Download (516) |
| SVP | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (629) |
| T | H9 | ENDODERM | GEO | Homo sapiens | GSE60606 | Download (23,419) | |
| T | H9 | MESODERM | GEO | Homo sapiens | GSE60606 | Download (33,640) | |
| TAF1 | GM12878 | ENCODE | Homo sapiens | ENCSR000BGS | Download (28,645) | ||
| TAF1 | GM12891 | ENCODE | Homo sapiens | ENCSR000BIM | Download (11,337) | ||
| TAF1 | GM12892 | ENCODE | Homo sapiens | ENCSR000BIB | Download (10,996) | ||
| TAF1 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BTO | Download (22,034) | ||
| TAF1 | liver | ENCODE | Homo sapiens | ENCSR016BMM | Download (13,676) | ||
| TAF1 | neural | ENCODE | Homo sapiens | ENCSR000BTX | Download (22,284) | ||
| TAF1 | PFSK1 | ENCODE | Homo sapiens | ENCSR000BQN | Download (10,236) | ||
| TAF1 | A-549 | ENCODE | Homo sapiens | ENCSR000BPF | Download (14,351) | ||
| TAF1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BJN | Download (17,810) | ||
| TAF1 | K-562 | ENCODE | Homo sapiens | ENCSR000BKS | Download (30,336) | ||
| TAF1 | MCF-7 | ENCODE | Homo sapiens | ENCSR000AHF | Download (7,388) | ||
| TAF1 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BQF | Download (41,497) | ||
| TAF1 | WA01 | ENCODE | Homo sapiens | ENCSR000BHO | Download (38,436) | ||
| TAF1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000BHT | Download (24,579) | ||
| TAF15 | Hep-G2 | ENCODE | Homo sapiens | ENCSR825MZS | Download (42,257) | ||
| TAF15 | K-562 | ENCODE | Homo sapiens | ENCSR047LSJ | Download (8,714) | ||
| TAF2 | hESC | GEO | Homo sapiens | GSE17917 | Download (1,561) | ||
| TAF3 | HCT-116 | GEO | Homo sapiens | GSE43539 | Download (21,122) | ||
| TAF7 | K-562 | ENCODE | Homo sapiens | ENCSR671GFC | Download (9,257) | ||
| TAF7 | WA01 | ENCODE | Homo sapiens | ENCSR000BLU | Download (10,955) | ||
| TAF7 | K-562 | ENCODE | Homo sapiens | ENCSR000BNM | Download (4,028) | ||
| TAF9B | K-562 | ENCODE | Homo sapiens | ENCSR100UQX | Download (6,464) | ||
| TAL1 | CHRF28811 | ENA | Homo sapiens | ERP008568 | Download (23,128) | ||
| TAL1 | K-562 | ENCODE | Homo sapiens | ENCSR000EHB | Download (33,885) | ||
| TAL1 | K-562 | ENCODE | Homo sapiens | ENCSR106FRG | Download (24,172) | ||
| TAL1 | ME-1 | GEO | Homo sapiens | GSE46044 | Download (22,032) | ||
| TAL1 | HSPC-CD34pos | GEO | Homo sapiens | GSE93372 | Download (17,524) | ||
| TAL1 | HSPC | GEO | Homo sapiens | GSE59657 | Download (9) | ||
| TAL1 | Jurkat | GEO | Homo sapiens | GSE25000 | Download (5,192) | ||
| TAL1 | Jurkat | GEO | Homo sapiens | GSE29180 | Download (3,761) | ||
| TAL1 | PRIMA2 | GEO | Homo sapiens | GSE33850 | Download (49,408) | ||
| TAL1 | PRIMA5 | GEO | Homo sapiens | GSE33850 | Download (48,683) | ||
| TAL1 | ProEs | GEO | Homo sapiens | GSE59087 | Download (36,669) | ||
| TAL1 | RPMI8402 | GEO | Homo sapiens | GSE39179 | Download (5,852) | ||
| TAL1 | CCRF-CEM | GEO | Homo sapiens | GSE33850 | Download (3,501) | ||
| TAL1 | MOLT-3 | GEO | Homo sapiens | GSE59657 | Download (37,959) | ||
| TAL1 | TSU-1621MT | GEO | Homo sapiens | GSE60477 | Download (31,488) | ||
| TAL1 | K-562 | GEO | Homo sapiens | GSE107726 | Download (43,054) | ||
| TAL1 | K-562 | MYO1D-Hub_KO | GEO | Homo sapiens | GSE107726 | Download (5,740) | |
| TAL1 | K-562 | MYO1D-Non-hub_KO | GEO | Homo sapiens | GSE107726 | Download (9,830) | |
| TAL1 | CD34 | GEO | Homo sapiens | GSE52924 | Download (28,594) | ||
| TAL1 | ECFC | GEO | Homo sapiens | GSE53423 | Download (82,261) | ||
| TAL1 | ECFC | TSA | GEO | Homo sapiens | GSE53423 | Download (96,261) | |
| TAL1 | erythroid | GEO | Homo sapiens | GSE25000 | Download (706) | ||
| TAL1 | erythroid | GEO | Homo sapiens | GSE42390 | Download (15,165) | ||
| TARDBP | MCF-7 | ENCODE | Homo sapiens | ENCSR801SWX | Download (17,150) | ||
| TARDBP | GM12878 | ENCODE | Homo sapiens | ENCSR016UEH | Download (8,314) | ||
| TARDBP | GM12878 | ENCODE | Homo sapiens | ENCSR412QBS | Download (71,929) | ||
| TARDBP | HEK293T | ENCODE | Homo sapiens | ENCSR753GIA | Download (1,223) | ||
| TARDBP | Hep-G2 | ENCODE | Homo sapiens | ENCSR145CXH | Download (23,983) | ||
| TARDBP | K-562 | ENCODE | Homo sapiens | ENCSR033VAZ | Download (9,121) | ||
| TARDBP | K-562 | ENCODE | Homo sapiens | ENCSR353HEP | Download (13,405) | ||
| TARDBP | K-562 | ENCODE | Homo sapiens | ENCSR429XTR | Download (33,355) | ||
| TASOR | HeLa | GEO | Homo sapiens | GSE95451 | Download (341) | ||
| TASOR | K-562 | MORC2-KO | GEO | Homo sapiens | GSE95374 | Download (108) | |
| TASOR | K-562 | MPP8-KO | GEO | Homo sapiens | GSE95374 | Download (120) | |
| TASOR | K-562 | TASOR-KO | GEO | Homo sapiens | GSE95374 | Download (203) | |
| TBL1X | HEK293T | GEO | Homo sapiens | GSE35197 | Download (43,682) | ||
| TBL1XR1 | GM12878 | ENCODE | Homo sapiens | ENCSR000DYZ | Download (2,148) | ||
| TBL1XR1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR101FJS | Download (33,234) | ||
| TBL1XR1 | K-562 | ENCODE | Homo sapiens | ENCSR000EGA | Download (9,053) | ||
| TBL1XR1 | K-562 | ENCODE | Homo sapiens | ENCSR000EGB | Download (18,786) | ||
| TBP | GM12878 | ENCODE | Homo sapiens | ENCSR000DZZ | Download (7,211) | ||
| TBP | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEL | Download (27,569) | ||
| TBP | K-562 | ENCODE | Homo sapiens | ENCSR000EHA | Download (27,416) | ||
| TBP | WA01 | ENCODE | Homo sapiens | ENCSR000ECB | Download (31,560) | ||
| TBP | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EDD | Download (31,393) | ||
| TBP | HBTEC | DM_MOI100_12hpi | GEO | Homo sapiens | GSE116772 | Download (705) | |
| TBP | HBTEC | DM_MOI100_18hpi | GEO | Homo sapiens | GSE116772 | Download (363) | |
| TBP | HBTEC | MOI20_12hpi | GEO | Homo sapiens | GSE116772 | Download (8,497) | |
| TBP | HBTEC | MOI20_18hpi | GEO | Homo sapiens | GSE116772 | Download (12,637) | |
| TBP | HBTEC | MOI5_dl312_18hpi | GEO | Homo sapiens | GSE116772 | Download (677) | |
| TBP | ME-1 | GEO | Homo sapiens | GSE46044 | Download (15,031) | ||
| TBP | K-562 | GEO | Homo sapiens | GSE55306 | Download (35,883) | ||
| TBX2 | Kelly | GEO | Homo sapiens | GSE94822 | Download (30,801) | ||
| TBX2 | SK-N-BE2-C | GEO | Homo sapiens | GSE94822 | Download (24,928) | ||
| TBX21 | CD4 | TH1 | GEO | Homo sapiens | GSE62482 | Download (21,384) | |
| TBX21 | GM12878 | ENCODE | Homo sapiens | ENCSR739IHN | Download (68,287) | ||
| TBX21 | TH1 | CD3-CD28_donor2 | GEO | Homo sapiens | GSE81881 | Download (54,917) | |
| TBX3 | Hep-G2 | ENCODE | Homo sapiens | ENCSR238QRG | Download (12,395) | ||
| TBX3 | Hep-G2 | ENCODE | Homo sapiens | ENCSR605YWG | Download (4,180) | ||
| TBX5 | cardiomyocyte | GEO | Homo sapiens | GSE85628 | Download (16,545) | ||
| TBX5 | cardiomyocyte | 1 | GEO | Homo sapiens | GSE85628 | Download (16,529) | |
| TBX5 | cardiomyocyte | 5 | GEO | Homo sapiens | GSE85628 | Download (21,863) | |
| TBX5 | cardiomyocyte | 7 | GEO | Homo sapiens | GSE85628 | Download (16,887) | |
| TBX5 | G296S | GEO | Homo sapiens | GSE85628 | Download (27,570) | ||
| TBX5 | G296S | 2 | GEO | Homo sapiens | GSE85628 | Download (27,552) | |
| TBX5 | G296S | 4 | GEO | Homo sapiens | GSE85628 | Download (31,201) | |
| TCF12 | ME-1 | GEO | Homo sapiens | GSE46044 | Download (27,543) | ||
| TCF12 | GM12878 | ENCODE | Homo sapiens | ENCSR000BGZ | Download (70,377) | ||
| TCF12 | GM12878 | ENCODE | Homo sapiens | ENCSR725VFL | Download (9,242) | ||
| TCF12 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BUV | Download (43,739) | ||
| TCF12 | A-549 | ENCODE | Homo sapiens | ENCSR000BQQ | Download (66,651) | ||
| TCF12 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BJG | Download (18,337) | ||
| TCF12 | K-562 | ENCODE | Homo sapiens | ENCSR189TRZ | Download (15,635) | ||
| TCF12 | K-562 | ENCODE | Homo sapiens | ENCSR744WOO | Download (26,968) | ||
| TCF12 | MCF-7 | ENCODE | Homo sapiens | ENCSR000BUN | Download (11,224) | ||
| TCF12 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BSL | Download (104,766) | ||
| TCF12 | WA01 | ENCODE | Homo sapiens | ENCSR000BIT | Download (42,144) | ||
| TCF12 | Jurkat | GEO | Homo sapiens | GSE29180 | Download (1,892) | ||
| TCF12 | RPMI8402 | GEO | Homo sapiens | GSE39179 | Download (5,425) | ||
| TCF12 | CCRF-CEM | GEO | Homo sapiens | GSE33850 | Download (831) | ||
| TCF12 | Kasumi-1 | GEO | Homo sapiens | GSE23730 | Download (39,275) | ||
| TCF12 | Kasumi-1 | GEO | Homo sapiens | GSE43834 | Download (19,422) | ||
| TCF25 | Hep-G2 | ENCODE | Homo sapiens | ENCSR110QXM | Download (8,030) | ||
| TCF3 | GM12878 | ENCODE | Homo sapiens | ENCSR000BQT | Download (62,865) | ||
| TCF3 | Jurkat | GEO | Homo sapiens | GSE29180 | Download (92) | ||
| TCF3 | RPMI8402 | GEO | Homo sapiens | GSE39179 | Download (1,528) | ||
| TCF3 | SEM | GEO | Homo sapiens | GSE85988 | Download (6,276) | ||
| TCF3 | CCRF-CEM | GEO | Homo sapiens | GSE33850 | Download (598) | ||
| TCF3 | Kasumi-1 | GEO | Homo sapiens | GSE43834 | Download (75,760) | ||
| TCF3 | RCH-ACV | GEO | Homo sapiens | GSE85988 | Download (50,197) | ||
| TCF4 | LS180 | GEO | Homo sapiens | GSE31939 | Download (8,271) | ||
| TCF4 | LS180 | 125 | GEO | Homo sapiens | GSE31939 | Download (9,537) | |
| TCF4 | CAL-1 | GEO | Homo sapiens | GSE76147 | Download (36,963) | ||
| TCF4 | GEN2-2 | GEO | Homo sapiens | GSE76147 | Download (54,638) | ||
| TCF4 | SH-SY5Y | GEO | Homo sapiens | GSE96915 | Download (10,912) | ||
| TCF7 | GM12878 | ENCODE | Homo sapiens | ENCSR501DKS | Download (10,895) | ||
| TCF7 | Hep-G2 | ENCODE | Homo sapiens | ENCSR444LIN | Download (22,878) | ||
| TCF7 | K-562 | ENCODE | Homo sapiens | ENCSR863KUB | Download (7,612) | ||
| TCF7L2 | K-562 | ENCODE | Homo sapiens | ENCSR888XZK | Download (4,496) | ||
| TCF7L2 | MCF-7 | ENCODE | Homo sapiens | ENCSR000EWT | Download (9,287) | ||
| TCF7L2 | PANC-1 | ENCODE | Homo sapiens | ENCSR000EXL | Download (26,149) | ||
| TCF7L2 | HCT-116 | ENCODE | Homo sapiens | ENCSR000EUV | Download (51,505) | ||
| TCF7L2 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EVE | Download (20,407) | ||
| TCF7L2 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EVF | Download (2,953) | ||
| TCF7L2 | CD34 | PROG_BIO | GEO | Homo sapiens | GSE29194 | Download (12,595) | |
| TCF7L2 | LNCaP | GEO | Homo sapiens | GSE51621 | Download (27,910) | ||
| TCF7L2 | MDA-MB-453 | GEO | Homo sapiens | GSE45201 | Download (19,570) | ||
| TCF7L2 | MDA-MB-453 | DHT | GEO | Homo sapiens | GSE45201 | Download (343) | |
| TCF7L2 | HEK293 | ENCODE | Homo sapiens | ENCSR000EUY | Download (9,553) | ||
| TCF7L2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EVQ | Download (2,122) | ||
| TCOF1 | HeLa | GEO | Homo sapiens | GSE89420 | Download (283) | ||
| TCP15 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (774) |
| TCP20 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (865) |
| TCP21 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,454) |
| TCP22 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,101) |
| TCP24 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (354) |
| TCP3 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (247) |
| TCP9 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (112) |
| TCX3 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,428) |
| TEAD1 | MCF-7 | GEO | Homo sapiens | GSE107013 | Download (93,503) | ||
| TEAD1 | T-47D | GEO | Homo sapiens | GSE107013 | Download (81,464) | ||
| TEAD1 | CCLP1 | GEO | Homo sapiens | GSE62272 | Download (7,669) | ||
| TEAD1 | pancreas | 12D | ENA | Homo sapiens | ERP008682 | Download (22,965) | |
| TEAD1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR497JLX | Download (100,070) | ||
| TEAD1 | HFOB | DIFF | GEO | Homo sapiens | GSE82295 | Download (1,100) | |
| TEAD1 | HUCCT1 | GEO | Homo sapiens | GSE68296 | Download (52,281) | ||
| TEAD1 | MSTO | GEO | Homo sapiens | GSE68170 | Download (29,852) | ||
| TEAD1 | H69 | GEO | Homo sapiens | GSE62274 | Download (34,737) | ||
| TEAD2 | K-562 | ENCODE | Homo sapiens | ENCSR635GTR | Download (1,484) | ||
| TEAD3 | Hep-G2 | ENCODE | Homo sapiens | ENCSR666QNP | Download (213,230) | ||
| TEAD4 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BSW | Download (53,703) | ||
| TEAD4 | A-549 | ENCODE | Homo sapiens | ENCSR000BUD | Download (19,912) | ||
| TEAD4 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BRP | Download (18,636) | ||
| TEAD4 | K-562 | ENCODE | Homo sapiens | ENCSR000BRK | Download (97,923) | ||
| TEAD4 | HUCCT1 | GEO | Homo sapiens | GSE68296 | Download (10,439) | ||
| TEAD4 | MKN28 | GEO | Homo sapiens | GSE44416 | Download (7,636) | ||
| TEAD4 | WA09 | DMSO | GEO | Homo sapiens | GSE52437 | Download (32) | |
| TEAD4 | MDA-MB-231 | GEO | Homo sapiens | GSE66081 | Download (20,276) | ||
| TEAD4 | SNU-216 | GEO | Homo sapiens | GSE44416 | Download (32,954) | ||
| TEAD4 | SK-N-BE2 | GEO | Homo sapiens | GSE84389 | Download (48,599) | ||
| TEAD4 | hESC | GEO | Homo sapiens | GSE99202 | Download (2,961) | ||
| TEAD4 | SK-MEL-147 | GEO | Homo sapiens | GSE94488 | Download (64,548) | ||
| TEAD4 | MCF-7 | ENCODE | Homo sapiens | ENCSR000BUO | Download (10,903) | ||
| TEAD4 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BUQ | Download (30,884) | ||
| TEAD4 | WA01 | ENCODE | Homo sapiens | ENCSR000BRY | Download (82,493) | ||
| TEAD4 | HCT-116 | ENCODE | Homo sapiens | ENCSR000BVJ | Download (7,349) | ||
| TEAD4 | BE2C | GEO | Homo sapiens | GSE84389 | Download (47,951) | ||
| TERF1 | LCL | GEO | Homo sapiens | GSE55053 | Download (39,652) | ||
| TERF2 | LCL | GEO | Homo sapiens | GSE55053 | Download (20,850) | ||
| TERF2 | myotube | TERF2_ovexp | GEO | Homo sapiens | GSE88983 | Download (123) | |
| TERF2 | HeLa | GEO | Homo sapiens | GSE46237 | Download (504) | ||
| TERF2 | WI-38VA13 | GEO | Homo sapiens | GSE46237 | Download (549) | ||
| TERT | hESC-1 | GEO | Homo sapiens | GSE77146 | Download (228) | ||
| TERT | BLM | GEO | Homo sapiens | GSE77146 | Download (257) | ||
| TERT | LOX-IMVI | GEO | Homo sapiens | GSE77146 | Download (216) | ||
| TERT | P493-6 | GEO | Homo sapiens | GSE77146 | Download (357) | ||
| TET2 | Jurkat | GEO | Homo sapiens | GSE85524 | Download (117,186) | ||
| TET2 | Jurkat | NCKD | GEO | Homo sapiens | GSE85524 | Download (8,372) | |
| TET2 | Jurkat | RUNX1KD | GEO | Homo sapiens | GSE85524 | Download (2,992) | |
| TET3 | HEK293T | GEO | Homo sapiens | GSE53809 | Download (2) | ||
| TFAP2A | WA09 | GEO | Homo sapiens | GSE105081 | Download (16,872) | ||
| TFAP2A | MCF-7 | GEO | Homo sapiens | GSE60270 | Download (9,954) | ||
| TFAP2A | MCF-7 | E2 | GEO | Homo sapiens | GSE60270 | Download (33,496) | |
| TFAP2C | SKBR3 | GEO | Homo sapiens | GSE36351 | Download (107,314) | ||
| TFAP2C | BT-474 | GEO | Homo sapiens | GSE36351 | Download (103,359) | ||
| TFAP2C | A-375 | DMSO | GEO | Homo sapiens | GSE41234 | Download (82,543) | |
| TFAP2C | MCF-7 | GEO | Homo sapiens | GSE21234 | Download (89,865) | ||
| TFAP2C | MCF-7 | E2 | GEO | Homo sapiens | GSE23852 | Download (44,597) | |
| TFAP2C | MCF-7 | ETOH | GEO | Homo sapiens | GSE23852 | Download (48,459) | |
| TFAP2C | MCF-7 | GEO | Homo sapiens | GSE36351 | Download (89,868) | ||
| TFAP2C | MDA-MB-453 | GEO | Homo sapiens | GSE36351 | Download (80,139) | ||
| TFAP2C | UCLA1-hESCs | 5iLAF-media | GEO | Homo sapiens | GSE101074 | Download (111,448) | |
| TFAP2C | UCLA1-hESCs | TFAP2C--_1d_TFAP2C_Induc | GEO | Homo sapiens | GSE101074 | Download (66,476) | |
| TFAP2C | UCLA1-hESCs | TFAP2C--_2d_TFAP2C_Induc | GEO | Homo sapiens | GSE101074 | Download (49,027) | |
| TFAP2C | WA09 | GEO | Homo sapiens | GSE105081 | Download (5,440) | ||
| TFAP4 | Hep-G2 | ENCODE | Homo sapiens | ENCSR103SZL | Download (44,862) | ||
| TFAP4 | LNCaP | GEO | Homo sapiens | GSE28857 | Download (13,236) | ||
| TFAP4 | DLD-1 | GEO | Homo sapiens | GSE46935 | Download (25,482) | ||
| TFAP4 | Kasumi-1 | GEO | Homo sapiens | GSE45738 | Download (15,893) | ||
| TFDP1 | K-562 | ENCODE | Homo sapiens | ENCSR017GBO | Download (6,193) | ||
| TFDP1 | K-562 | ENCODE | Homo sapiens | ENCSR224IKA | Download (13,152) | ||
| TFDP1 | MM1-S | GEO | Homo sapiens | GSE80661 | Download (8,899) | ||
| TFDP1 | U266 | GEO | Homo sapiens | GSE80661 | Download (2,873) | ||
| TFE3 | Hep-G2 | ENCODE | Homo sapiens | ENCSR589SNT | Download (71,138) | ||
| TFIIIC | HEK293 | GEO | Homo sapiens | GSE119418 | Download (25,289) | ||
| TFL2 | Col-0 | Col-0_seedling | 14 | GEO | Arabidopsis thaliana | GSE76571 | Download (11,703) |
| TGA10 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (6,140) |
| TGA2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (618) |
| TGA3 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (540) |
| TGA4 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,079) |
| TGA4 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (6,484) |
| TGA5 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,551) |
| TGA6 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,940) |
| TGA7 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,019) |
| TGA9 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,490) |
| TGIF2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR993LMB | Download (15,394) | ||
| THAP1 | K-562 | ENCODE | Homo sapiens | ENCSR000BNN | Download (33,325) | ||
| THAP11 | Hep-G2 | ENCODE | Homo sapiens | ENCSR562POI | Download (90,253) | ||
| THAP11 | HeLa | GEO | Homo sapiens | GSE31417 | Download (423) | ||
| THRA | K-562 | ENCODE | Homo sapiens | ENCSR264CZJ | Download (836) | ||
| THRAP3 | K-562 | ENCODE | Homo sapiens | ENCSR871TKJ | Download (4,284) | ||
| THRB | Hep-G2 | ENCODE | Homo sapiens | ENCSR430JGJ | Download (43,669) | ||
| TINY | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,729) |
| TLE3 | LNCaP | GEO | Homo sapiens | GSE94682 | Download (70,331) | ||
| TLE3 | LNCaP | r1881 | GEO | Homo sapiens | GSE94682 | Download (61,506) | |
| TLE3 | MCF-7 | GEO | Homo sapiens | GSE41561 | Download (7,873) | ||
| TOP1 | LNCaP | GEO | Homo sapiens | GSE63202 | Download (5,244) | ||
| TOP1 | LNCaP | DHT | GEO | Homo sapiens | GSE63202 | Download (8,584) | |
| TOP1 | HCT-116 | JQ1 | GEO | Homo sapiens | GSE57628 | Download (180) | |
| TOP1ALPHA | Ler | Ler_inflorescence | GEO | Arabidopsis thaliana | GSE53950 | Download (271) | |
| TOP2A | K-562 | Etoposide | GEO | Homo sapiens | GSE79593 | Download (146) | |
| TOP2A | K-562 | Genistein | GEO | Homo sapiens | GSE79593 | Download (158,766) | |
| TOP2A | K-562 | Mitoxantrone | GEO | Homo sapiens | GSE79593 | Download (172,266) | |
| TOP2A | K-562 | pBQ | GEO | Homo sapiens | GSE79593 | Download (830) | |
| TP53 | hESC | DAMAGED | GEO | Homo sapiens | GSE39912 | Download (19,584) | |
| TP53 | hESC | DIFF | GEO | Homo sapiens | GSE39912 | Download (15,054) | |
| TP53 | keratinocyte | GEO | Homo sapiens | GSE56674 | Download (4,636) | ||
| TP53 | keratinocyte | ADRIA | GEO | Homo sapiens | GSE56674 | Download (7,891) | |
| TP53 | keratinocyte | CISP | GEO | Homo sapiens | GSE56674 | Download (14,925) | |
| TP53 | lymphocyte | 104_Nutlin | GEO | Homo sapiens | GSE110368 | Download (1,715) | |
| TP53 | lymphocyte | 116 | GEO | Homo sapiens | GSE110368 | Download (989) | |
| TP53 | lymphocyte | 116_DXR | GEO | Homo sapiens | GSE110368 | Download (2,028) | |
| TP53 | lymphocyte | 116_Nutlin | GEO | Homo sapiens | GSE110368 | Download (8,180) | |
| TP53 | lymphocyte | 45 | GEO | Homo sapiens | GSE110368 | Download (531) | |
| TP53 | lymphocyte | 45_DXR | GEO | Homo sapiens | GSE110368 | Download (2,947) | |
| TP53 | lymphocyte | 45_Nutlin | GEO | Homo sapiens | GSE110368 | Download (1,545) | |
| TP53 | lymphocyte | 90 | GEO | Homo sapiens | GSE110368 | Download (7,275) | |
| TP53 | lymphocyte | 90_DXR | GEO | Homo sapiens | GSE110368 | Download (2,744) | |
| TP53 | lymphocyte | 90_Nutlin | GEO | Homo sapiens | GSE110368 | Download (1,508) | |
| TP53 | SW480 | shLacZ_16h_TNF-a | GEO | Homo sapiens | GSE115985 | Download (108,925) | |
| TP53 | SW480 | shp53_16h_TNF-a | GEO | Homo sapiens | GSE115985 | Download (35,985) | |
| TP53 | SW480 | GEO | Homo sapiens | GSE67108 | Download (12) | ||
| TP53 | U2OS | 10Gy_2h_recovery | GEO | Homo sapiens | GSE110800 | Download (170) | |
| TP53 | U2OS | 10Gy_4h_recovery | GEO | Homo sapiens | GSE110800 | Download (227) | |
| TP53 | U2OS | ACTD | GEO | Homo sapiens | GSE21939 | Download (522) | |
| TP53 | U2OS | ETO | GEO | Homo sapiens | GSE21939 | Download (598) | |
| TP53 | U2OS | ACTD | GEO | Homo sapiens | GSE21939 | Download (84) | |
| TP53 | U2OS | ETO | GEO | Homo sapiens | GSE21939 | Download (125) | |
| TP53 | U2OS | ACTD | GEO | Homo sapiens | GSE21939 | Download (550) | |
| TP53 | U2OS | ETO | GEO | Homo sapiens | GSE21939 | Download (1,103) | |
| TP53 | U2OS | GEO | Homo sapiens | GSE46641 | Download (608) | ||
| TP53 | U2OS | DMSO | GEO | Homo sapiens | GSE46641 | Download (3,234) | |
| TP53 | U2OS | DXR | GEO | Homo sapiens | GSE46641 | Download (2,397) | |
| TP53 | U2OS | NUT | GEO | Homo sapiens | GSE46641 | Download (16,966) | |
| TP53 | H9 | GEO | Homo sapiens | GSE39912 | Download (2,839) | ||
| TP53 | Hep-G2 | GEO | Homo sapiens | GSE64877 | Download (245) | ||
| TP53 | IMR-90 | GEO | Homo sapiens | GSE115940 | Download (4,587) | ||
| TP53 | IMR-90 | GEO | Homo sapiens | GSE31558 | Download (1,145) | ||
| TP53 | IMR-90 | GEO | Homo sapiens | GSE42728 | Download (9,865) | ||
| TP53 | IMR-90 | SENES_SHLUC | GEO | Homo sapiens | GSE42728 | Download (19,659) | |
| TP53 | IMR-90 | SENES_SHP53 | GEO | Homo sapiens | GSE42728 | Download (731) | |
| TP53 | IMR-90 | APO | GEO | Homo sapiens | GSE53491 | Download (23,452) | |
| TP53 | IMR-90 | SENE | GEO | Homo sapiens | GSE53491 | Download (5,426) | |
| TP53 | IMR-90 | DMSO | GEO | Homo sapiens | GSE58740 | Download (354) | |
| TP53 | IMR-90 | NUT3A | GEO | Homo sapiens | GSE58740 | Download (7,809) | |
| TP53 | MCF-7 | NUT | GEO | Homo sapiens | GSE47041 | Download (209) | |
| TP53 | MCF-7 | nutlin | GEO | Homo sapiens | GSE86164 | Download (34,147) | |
| TP53 | SaOS-2 | GEO | Homo sapiens | GSE15780 | Download (28,570) | ||
| TP53 | SaOS-2 | GEO | Homo sapiens | GSE51268 | Download (10,885) | ||
| TP53 | SJSA-1 | GEO | Homo sapiens | GSE86164 | Download (2,028) | ||
| TP53 | SJSA-1 | nutlin | GEO | Homo sapiens | GSE86164 | Download (10,231) | |
| TP53 | HCT-116 | GEO | Homo sapiens | GSE58506 | Download (666) | ||
| TP53 | HCT-116 | 5FU | GEO | Homo sapiens | GSE58506 | Download (3,383) | |
| TP53 | HCT-116 | GEO | Homo sapiens | GSE58714 | Download (63) | ||
| TP53 | HCT-116 | IR | GEO | Homo sapiens | GSE60267 | Download (7,301) | |
| TP53 | HCT-116 | CAMP | GEO | Homo sapiens | GSE74355 | Download (82) | |
| TP53 | HCT-116 | DMSO_KOATF3 | GEO | Homo sapiens | GSE74355 | Download (1,049) | |
| TP53 | HCT-116 | nutlin | GEO | Homo sapiens | GSE86164 | Download (5,944) | |
| TP53 | MDA-MB-231 | GEO | Homo sapiens | GSE95303 | Download (67,603) | ||
| TP53 | BC3 | ENA | Homo sapiens | ERP014035 | Download (87) | ||
| TP53 | BC3 | NUT | ENA | Homo sapiens | ERP014035 | Download (1,165) | |
| TP53 | BC3 | TPA_24H | ENA | Homo sapiens | ERP014035 | Download (146) | |
| TP53 | BC3 | TPA_4H | ENA | Homo sapiens | ERP014035 | Download (44) | |
| TP53 | SW480 | 0h_TNFa | GEO | Homo sapiens | GSE102796 | Download (48,447) | |
| TP53 | SW480 | 16h_TNFa | GEO | Homo sapiens | GSE102796 | Download (102,210) | |
| TP53 | U2OS | ENA | Homo sapiens | ERP004176 | Download (1,024) | ||
| TP53 | U2OS | UV_16H | ENA | Homo sapiens | ERP004176 | Download (2,061) | |
| TP53 | U2OS | UV_8H | ENA | Homo sapiens | ERP004176 | Download (2,192) | |
| TP53 | U2OS | 2h_4GY | GEO | Homo sapiens | GSE100292 | Download (253) | |
| TP53 | A-549 | 2h_4GY | GEO | Homo sapiens | GSE100292 | Download (2,278) | |
| TP53 | A-498 | 2h_4GY | GEO | Homo sapiens | GSE100292 | Download (1,824) | |
| TP53 | LOX-IMVI | 2h_4GY | GEO | Homo sapiens | GSE100292 | Download (941) | |
| TP53 | MALME-3 | 2h_4GY | GEO | Homo sapiens | GSE100292 | Download (103) | |
| TP53 | MCF-7 | 1h_IR_10Gy | GEO | Homo sapiens | GSE100099 | Download (3,912) | |
| TP53 | MCF-7 | 2-5h_IR_10Gy | GEO | Homo sapiens | GSE100099 | Download (5,979) | |
| TP53 | MCF-7 | 4h_IR_10Gy | GEO | Homo sapiens | GSE100099 | Download (3,190) | |
| TP53 | MCF-7 | 7-5h_IR_10Gy | GEO | Homo sapiens | GSE100099 | Download (5,096) | |
| TP53 | MCF-7 | 7-5h_IR_10Gy_Nutlin | GEO | Homo sapiens | GSE100099 | Download (22,476) | |
| TP53 | MCF-7 | 2h_4GY | GEO | Homo sapiens | GSE100292 | Download (248) | |
| TP53 | MCF-7 | minus_Decitabine | GEO | Homo sapiens | GSE100292 | Download (468) | |
| TP53 | MCF-7 | nutlin_2h | GEO | Homo sapiens | GSE100292 | Download (1,289) | |
| TP53 | MCF-7 | plus_Decitabine | GEO | Homo sapiens | GSE100292 | Download (523) | |
| TP53 | MCF-7 | GEO | Homo sapiens | GSE101737 | Download (163) | ||
| TP53 | MCF-7 | NCS-treated | GEO | Homo sapiens | GSE101737 | Download (2,899) | |
| TP53 | NCI-H1299 | ENA | Homo sapiens | ERP022459 | Download (269) | ||
| TP53 | NCI-H460 | 2h_4GY | GEO | Homo sapiens | GSE100292 | Download (1,191) | |
| TP53 | SaOS-2 | ENA | Homo sapiens | ERP002038 | Download (1,553) | ||
| TP53 | SK-MEL-5 | 2h_4GY | GEO | Homo sapiens | GSE100292 | Download (104) | |
| TP53 | UACC-257 | 2h_4GY | GEO | Homo sapiens | GSE100292 | Download (1,896) | |
| TP53 | UACC-257 | nutlin_2h | GEO | Homo sapiens | GSE100292 | Download (538) | |
| TP53 | UACC-62 | 2h_4GY | GEO | Homo sapiens | GSE100292 | Download (138) | |
| TP53 | UO-31 | 2h_4GY | GEO | Homo sapiens | GSE100292 | Download (3,457) | |
| TP53 | HCT-116 | 2h_4GY | GEO | Homo sapiens | GSE100292 | Download (1,096) | |
| TP53 | RPE | 2h_4GY | GEO | Homo sapiens | GSE100292 | Download (343) | |
| TP53 | GM00011 | GEO | Homo sapiens | GSE55727 | Download (11,500) | ||
| TP53 | GM06170 | GEO | Homo sapiens | GSE55727 | Download (18,223) | ||
| TP63 | TE-5 | GEO | Homo sapiens | GSE106563 | Download (1,223) | ||
| TP63 | EP156T | GEO | Homo sapiens | GSE43111 | Download (4,809) | ||
| TP63 | HaCaT | caRAS_TGFB | GEO | Homo sapiens | GSE60814 | Download (14,079) | |
| TP63 | HaCaT | dnRAS_TGFB | GEO | Homo sapiens | GSE60814 | Download (8,126) | |
| TP63 | HaCaT | LacZ | GEO | Homo sapiens | GSE60814 | Download (2,936) | |
| TP63 | HaCaT | LacZ_TGFB | GEO | Homo sapiens | GSE60814 | Download (13,630) | |
| TP63 | HCC95 | GEO | Homo sapiens | GSE46837 | Download (3,158) | ||
| TP63 | keratinocyte | GEO | Homo sapiens | GSE33571 | Download (41,268) | ||
| TP63 | keratinocyte | diff | GEO | Homo sapiens | GSE33571 | Download (67,437) | |
| TP63 | keratinocyte | GEO | Homo sapiens | GSE56674 | Download (30,106) | ||
| TP63 | keratinocyte | ADRIA | GEO | Homo sapiens | GSE56674 | Download (4,194) | |
| TP63 | keratinocyte | CISP | GEO | Homo sapiens | GSE56674 | Download (27,804) | |
| TP63 | keratinocyte | D0 | GEO | Homo sapiens | GSE59824 | Download (46,719) | |
| TP63 | keratinocyte | D2 | GEO | Homo sapiens | GSE59824 | Download (53,232) | |
| TP63 | keratinocyte | D4 | GEO | Homo sapiens | GSE59824 | Download (48,524) | |
| TP63 | keratinocyte | D7 | GEO | Homo sapiens | GSE59824 | Download (44,355) | |
| TP63 | keratinocyte | epidermal | GEO | Homo sapiens | GSE67382 | Download (26,801) | |
| TP63 | keratinocyte | epidermal_KDPAF | GEO | Homo sapiens | GSE67382 | Download (14,350) | |
| TP63 | KYSE70 | GEO | Homo sapiens | GSE46837 | Download (2,868) | ||
| TP63 | TT | GEO | Homo sapiens | GSE46837 | Download (6,456) | ||
| TP63 | JHU-029 | GEO | Homo sapiens | GSE88859 | Download (15,134) | ||
| TP63 | MCF-10A | DCIS | GEO | Homo sapiens | GSE72009 | Download (15,364) | |
| TP63 | MDA-MB-231 | GEO | Homo sapiens | GSE72009 | Download (175) | ||
| TP73 | SaOS-2 | GEO | Homo sapiens | GSE15780 | Download (23,713) | ||
| TP73 | SaOS-2 | GEO | Homo sapiens | GSE15780 | Download (15,534) | ||
| TP73 | GM12878 | GEO | Homo sapiens | GSE97661 | Download (1,370) | ||
| TRB1 | Col-0 | Col-0_seedling | TRB1_14d | GEO | Arabidopsis thaliana | GSE69431 | Download (6,928) |
| TRB1 | Col-0 | Col-0_seedling | TRB1-GFP_14d | GEO | Arabidopsis thaliana | GSE69431 | Download (14,850) |
| TRB2 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,975) |
| TRIM22 | MCF-7 | ENCODE | Homo sapiens | ENCSR875PEI | Download (26,494) | ||
| TRIM22 | GM12878 | ENCODE | Homo sapiens | ENCSR637QAM | Download (35,983) | ||
| TRIM22 | GM12878 | ENCODE | Homo sapiens | ENCSR835XKS | Download (66,194) | ||
| TRIM22 | Hep-G2 | ENCODE | Homo sapiens | ENCSR506HXN | Download (2,486) | ||
| TRIM24 | LNCaP | DHT | GEO | Homo sapiens | GSE69331 | Download (10,714) | |
| TRIM24 | LNCaP | ETOH | GEO | Homo sapiens | GSE69331 | Download (14,663) | |
| TRIM24 | LNCaP-abl | ETOH | GEO | Homo sapiens | GSE69331 | Download (22,611) | |
| TRIM24 | K-562 | ENCODE | Homo sapiens | ENCSR907MZR | Download (46,671) | ||
| TRIM24 | K-562 | ENCODE | Homo sapiens | ENCSR957LDM | Download (22,490) | ||
| TRIM24 | MOLM-13 | dTRIM24 | GEO | Homo sapiens | GSE100571 | Download (41,552) | |
| TRIM24 | MOLM-13 | IACS-9571 | GEO | Homo sapiens | GSE100571 | Download (58,342) | |
| TRIM25 | BT-549 | GEO | Homo sapiens | GSE79588 | Download (31,403) | ||
| TRIM25 | MDA-MB-231 | GEO | Homo sapiens | GSE79588 | Download (16,379) | ||
| TRIM28 | TB40E | GEO | Homo sapiens | GSE53271 | Download (220,591) | ||
| TRIM28 | WIBR3 | GEO | Homo sapiens | GSE84382 | Download (2,922) | ||
| TRIM28 | WIBR3 | NAIVE | GEO | Homo sapiens | GSE84382 | Download (39,840) | |
| TRIM28 | WA01 | GEO | Homo sapiens | GSE78099 | Download (3,423) | ||
| TRIM28 | HEK293 | ENCODE | Homo sapiens | ENCSR000EUZ | Download (13,584) | ||
| TRIM28 | HEK293 | ENCODE | Homo sapiens | ENCSR618HNF | Download (17,859) | ||
| TRIM28 | U2OS | ENCODE | Homo sapiens | ENCSR000EYC | Download (18,412) | ||
| TRIM28 | K-562 | ENCODE | Homo sapiens | ENCSR000BRW | Download (23,905) | ||
| TRIM28 | K-562 | ENCODE | Homo sapiens | ENCSR000EVY | Download (7,786) | ||
| TRIM28 | K-562 | ENCODE | Homo sapiens | ENCSR474CVP | Download (21,766) | ||
| TRIM28 | HCT-116 | GEO | Homo sapiens | GSE72622 | Download (22,027) | ||
| TRIM28 | AF22 | GEO | Homo sapiens | GSE84259 | Download (39,913) | ||
| TRIP13 | K-562 | ENCODE | Homo sapiens | ENCSR154EIH | Download (1,264) | ||
| TRP1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (169) |
| TRP2 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (1,682) |
| TRP47 | THP-1 | GEO | Homo sapiens | GSE118453 | Download (129,288) | ||
| TRPS1 | MCF-7 | GEO | Homo sapiens | GSE107013 | Download (4,181) | ||
| TRPS1 | T-47D | GEO | Homo sapiens | GSE107013 | Download (20,767) | ||
| TRRAP | HeLa | GEO | Homo sapiens | GSE20303 | Download (137) | ||
| TSC22D4 | K-562 | ENCODE | Homo sapiens | ENCSR787RVK | Download (243) | ||
| TSC22D4 | GM12878 | GEO | Homo sapiens | GSE97661 | Download (3,714) | ||
| TSC22D4 | Hep-G2 | GEO | Homo sapiens | GSE97661 | Download (6,422) | ||
| TSHZ1 | HEK293 | ENCODE | Homo sapiens | ENCSR217WRC | Download (11,240) | ||
| TWIST1 | SHEP-21N | GEO | Homo sapiens | GSE80151 | Download (52,965) | ||
| TWIST1 | SHEP-21N | 24h | GEO | Homo sapiens | GSE80151 | Download (21,874) | |
| TWIST1 | SHEP-21N | DOX_0H | GEO | Homo sapiens | GSE80151 | Download (25,334) | |
| TWIST1 | SHEP-21N | DOX_24H | GEO | Homo sapiens | GSE80151 | Download (3,061) | |
| TWIST1 | SK-N-BE2-C | GEO | Homo sapiens | GSE80151 | Download (50,372) | ||
| TWIST1 | BE2C | GEO | Homo sapiens | GSE80151 | Download (49,172) | ||
| U2AF1 | K-562 | ENCODE | Homo sapiens | ENCSR690GUG | Download (4,325) | ||
| U2AF1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR868JLS | Download (27,283) | ||
| U2AF2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR991ADX | Download (5,923) | ||
| UBN1 | HeLa | GEO | Homo sapiens | GSE45024 | Download (24,725) | ||
| UBTF | HeLa-S3 | ENCODE | Homo sapiens | ENCSR634ZGP | Download (280) | ||
| UBTF | GM12878 | ENCODE | Homo sapiens | ENCSR459FTB | Download (5,596) | ||
| UBTF | K-562 | ENCODE | Homo sapiens | ENCSR000EFW | Download (13,835) | ||
| UBTF | K-562 | ENCODE | Homo sapiens | ENCSR000EFZ | Download (37,036) | ||
| UBTF | hESC | GEO | Homo sapiens | GSE76586 | Download (390) | ||
| UBTF | hESC | activinA | GEO | Homo sapiens | GSE76586 | Download (261) | |
| USF1 | GM12878 | ENCODE | Homo sapiens | ENCSR000BGI | Download (9,331) | ||
| USF1 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BSX | Download (32,021) | ||
| USF1 | A-549 | ENCODE | Homo sapiens | ENCSR000BHX | Download (10,399) | ||
| USF1 | A-549 | ENCODE | Homo sapiens | ENCSR000BJB | Download (14,479) | ||
| USF1 | A-549 | ENCODE | Homo sapiens | ENCSR000BPV | Download (17,749) | ||
| USF1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BGM | Download (28,325) | ||
| USF1 | K-562 | ENCODE | Homo sapiens | ENCSR000BKT | Download (43,123) | ||
| USF1 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BMF | Download (55,214) | ||
| USF1 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BTZ | Download (12,911) | ||
| USF1 | WA01 | ENCODE | Homo sapiens | ENCSR000BIU | Download (59,924) | ||
| USF1 | HCT-116 | ENCODE | Homo sapiens | ENCSR000BVK | Download (12,126) | ||
| USF2 | SK-N-SH | ENCODE | Homo sapiens | ENCSR945NFL | Download (3,234) | ||
| USF2 | WA01 | ENCODE | Homo sapiens | ENCSR000ECD | Download (8,530) | ||
| USF2 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECW | Download (25,222) | ||
| USF2 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZU | Download (4,484) | ||
| USF2 | A-549 | ENCODE | Homo sapiens | ENCSR563FBT | Download (9,342) | ||
| USF2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000EEF | Download (8,643) | ||
| USF2 | IMR-90 | ENCODE | Homo sapiens | ENCSR513UQG | Download (40,117) | ||
| USF2 | K-562 | ENCODE | Homo sapiens | ENCSR000EHG | Download (3,621) | ||
| USF2 | K-562 | ENCODE | Homo sapiens | ENCSR359NFW | Download (8,520) | ||
| USF2 | K-562 | GEO | Homo sapiens | GSE111469 | Download (17,186) | ||
| USF2 | GM12878 | GEO | Homo sapiens | GSE97661 | Download (34,608) | ||
| USF2 | Hep-G2 | GEO | Homo sapiens | GSE97661 | Download (21,811) | ||
| USP7 | HEK293T | GEO | Homo sapiens | GSE61048 | Download (8,855) | ||
| UTX | MCF-7 | GEO | Homo sapiens | GSE96996 | Download (477) | ||
| VDR | LCLGM10861 | CALCITRIOL | GEO | Homo sapiens | GSE22484 | Download (6,003) | |
| VDR | LNCaP | GEO | Homo sapiens | GSE64656 | Download (21,327) | ||
| VDR | LS180 | GEO | Homo sapiens | GSE31939 | Download (1,173) | ||
| VDR | LS180 | 125 | GEO | Homo sapiens | GSE31939 | Download (6,130) | |
| VDR | LX2 | GEO | Homo sapiens | GSE38103 | Download (2,449) | ||
| VDR | LX2 | CALCIPOTRIOL | GEO | Homo sapiens | GSE38103 | Download (2,065) | |
| VDR | LX2 | CALCIPOTRIOL_TGFB1 | GEO | Homo sapiens | GSE38103 | Download (28,598) | |
| VDR | THP-1 | GEO | Homo sapiens | GSE51181 | Download (9) | ||
| VDR | THP-1 | 1H_125D | GEO | Homo sapiens | GSE51181 | Download (170) | |
| VDR | THP-1 | 2H_125D | GEO | Homo sapiens | GSE51181 | Download (249) | |
| VDR | THP-1 | GEO | Homo sapiens | GSE53041 | Download (316) | ||
| VDR | THP-1 | CAL | GEO | Homo sapiens | GSE53041 | Download (1,039) | |
| VDR | THP-1 | 1d_1-25-OH-2D3 | GEO | Homo sapiens | GSE89431 | Download (57,025) | |
| VDR | THP-1 | 2h_1-25-OH-2D3 | GEO | Homo sapiens | GSE89431 | Download (3,720) | |
| VDR | THP-1 | EtOH_1d | GEO | Homo sapiens | GSE89431 | Download (8,672) | |
| VDR | THP-1 | EtOH_2h | GEO | Homo sapiens | GSE89431 | Download (1,307) | |
| VEZF1 | K-562 | ENCODE | Homo sapiens | ENCSR189YMA | Download (23,329) | ||
| VRN1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,578) |
| WDHD1 | MCF-7 | Ab_R1251-1-1A5 | GEO | Homo sapiens | GSE97661 | Download (5,006) | |
| WDHD1 | MCF-7 | Ab_R1251-1-1B10 | GEO | Homo sapiens | GSE97661 | Download (5,737) | |
| WDR5 | RS4-11 | GEO | Homo sapiens | GSE72864 | Download (855) | ||
| WIP5 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (445) |
| WRKY11 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (129) |
| WRKY14 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (750) |
| WRKY15 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,363) |
| WRKY18 | Col-0 | Col-0_seedling | 12d | GEO | Arabidopsis thaliana | GSE85922 | Download (16,559) |
| WRKY18 | Col-0 | Col-0_seedling | 12d-flg22 | GEO | Arabidopsis thaliana | GSE85922 | Download (13,718) |
| WRKY18 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,564) |
| WRKY18-33-40 | Col-0 | Col-0_seedling | 12d-mutant-flg22-0h | GEO | Arabidopsis thaliana | GSE109149 | Download (15,277) |
| WRKY18-33-40 | Col-0 | Col-0_seedling | 12d-mutant-flg22-2h | GEO | Arabidopsis thaliana | GSE109149 | Download (14,869) |
| WRKY18-33-40 | Col-0 | Col-0_seedling | 12d-wt-flg22-0h | GEO | Arabidopsis thaliana | GSE109149 | Download (14,542) |
| WRKY18-33-40 | Col-0 | Col-0_seedling | 12d-wt-flg22-2h | GEO | Arabidopsis thaliana | GSE109149 | Download (15,832) |
| WRKY20 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (497) |
| WRKY21 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (162) |
| WRKY22 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,260) |
| WRKY24 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (477) |
| WRKY25 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,395) |
| WRKY27 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (347) |
| WRKY28 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,101) |
| WRKY29 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (607) |
| WRKY3 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,179) |
| WRKY30 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (2,310) |
| WRKY31 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (257) |
| WRKY33 | Col-0 | Col-0_seedling | 12d | GEO | Arabidopsis thaliana | GSE85922 | Download (9,828) |
| WRKY33 | Col-0 | Col-0_seedling | 12d-flg22 | GEO | Arabidopsis thaliana | GSE85922 | Download (17,051) |
| WRKY33 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,676) |
| WRKY33 | Col-0 | Col-0_leaves | 4w-Bc | GEO | Arabidopsis thaliana | GSE66289 | Download (14,506) |
| WRKY33 | Col-0 | Col-0_leaves | 4w-mock | GEO | Arabidopsis thaliana | GSE66289 | Download (1,078) |
| WRKY40 | Col-0 | Col-0_seedling | 12d | GEO | Arabidopsis thaliana | GSE85922 | Download (6,438) |
| WRKY40 | Col-0 | Col-0_seedling | 12d-flg22 | GEO | Arabidopsis thaliana | GSE85922 | Download (13,849) |
| WRKY45 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,814) |
| WRKY50 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (651) |
| WRKY6 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (310) |
| WRKY65 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (385) |
| WRKY70 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,721) |
| WRKY71 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (3,159) |
| WRKY8 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (795) |
| WRNIP1 | GM12878 | ENCODE | Homo sapiens | ENCSR000EAA | Download (67) | ||
| WT1 | HEK293 | ENCODE | Homo sapiens | ENCSR966PJJ | Download (40,051) | ||
| WT1 | K-562 | streptavidin | GEO | Homo sapiens | GSE81009 | Download (107,206) | |
| WT1 | K-562 | streptavidin | GEO | Homo sapiens | GSE81009 | Download (348,909) | |
| XBP1 | HS578T | HYPO_GLUDEP | GEO | Homo sapiens | GSE49952 | Download (3,929) | |
| XBP1 | MDA-MB-231 | GEO | Homo sapiens | GSE49952 | Download (216,881) | ||
| XBP1 | MDA-MB-231 | HYPO_GLUDEP | GEO | Homo sapiens | GSE49952 | Download (256,241) | |
| XBP1 | T-47D | HYPO_GLUDEP | GEO | Homo sapiens | GSE49952 | Download (85) | |
| XRCC3 | K-562 | ENCODE | Homo sapiens | ENCSR997NGQ | Download (600) | ||
| XRCC5 | Hep-G2 | ENCODE | Homo sapiens | ENCSR031URL | Download (172,890) | ||
| XRCC5 | K-562 | ENCODE | Homo sapiens | ENCSR506KWJ | Download (48,448) | ||
| YAP1 | MCF-7 | GEO | Homo sapiens | GSE107013 | Download (9,114) | ||
| YAP1 | CCLP1 | GEO | Homo sapiens | GSE62272 | Download (91) | ||
| YAP1 | HUCCT1 | GEO | Homo sapiens | GSE68296 | Download (167) | ||
| YAP1 | MSTO | GEO | Homo sapiens | GSE68170 | Download (1,199) | ||
| YAP1 | H69 | GEO | Homo sapiens | GSE62274 | Download (1,175) | ||
| YAP1 | MDA-MB-231 | GEO | Homo sapiens | GSE66081 | Download (15,906) | ||
| YAP1 | OVCAR-5 | GEO | Homo sapiens | GSE57236 | Download (8,916) | ||
| YAP1 | MCF-10A | GEO | Homo sapiens | GSE97972 | Download (12,857) | ||
| YAP1 | WA01 | GEO | Homo sapiens | GSE99202 | Download (646) | ||
| YBX1 | MCF-7 | ENCODE | Homo sapiens | ENCSR864KNH | Download (970) | ||
| YBX1 | GM12878 | ENCODE | Homo sapiens | ENCSR205SKQ | Download (3,125) | ||
| YBX1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR799GOY | Download (4,914) | ||
| YBX1 | K-562 | ENCODE | Homo sapiens | ENCSR107RHZ | Download (1,696) | ||
| YBX3 | K-562 | ENCODE | Homo sapiens | ENCSR567JEU | Download (2,003) | ||
| YY1 | GM12878 | ENCODE | Homo sapiens | ENCSR000BNP | Download (94,804) | ||
| YY1 | GM12878 | ENCODE | Homo sapiens | ENCSR000EUM | Download (987) | ||
| YY1 | GM12891 | ENCODE | Homo sapiens | ENCSR000BKJ | Download (38,612) | ||
| YY1 | GM12892 | ENCODE | Homo sapiens | ENCSR000BLT | Download (45,301) | ||
| YY1 | HEK293 | ENCODE | Homo sapiens | ENCSR859RAO | Download (10,454) | ||
| YY1 | Ishikawa | ENCODE | Homo sapiens | ENCSR000BSY | Download (45,378) | ||
| YY1 | liver | ENCODE | Homo sapiens | ENCSR382MOM | Download (82,554) | ||
| YY1 | liver | ENCODE | Homo sapiens | ENCSR994YLZ | Download (49,120) | ||
| YY1 | A-549 | ENCODE | Homo sapiens | ENCSR000BPM | Download (35,915) | ||
| YY1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BNT | Download (40,358) | ||
| YY1 | K-562 | ENCODE | Homo sapiens | ENCSR000BKU | Download (44,773) | ||
| YY1 | K-562 | ENCODE | Homo sapiens | ENCSR000BMH | Download (59,520) | ||
| YY1 | K-562 | ENCODE | Homo sapiens | ENCSR000EWF | Download (10,973) | ||
| YY1 | NT2-D1 | ENCODE | Homo sapiens | ENCSR000EXG | Download (15,117) | ||
| YY1 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BLZ | Download (32,832) | ||
| YY1 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BSM | Download (22,973) | ||
| YY1 | WA01 | ENCODE | Homo sapiens | ENCSR000BKD | Download (68,977) | ||
| YY1 | HCT-116 | ENCODE | Homo sapiens | ENCSR000BNX | Download (53,424) | ||
| YY1 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (116,981) | ||
| YY1 | HeLa | GEO | Homo sapiens | GSE31417 | Download (1,058) | ||
| YY1 | AB-LCL | GEO | Homo sapiens | GSE98477 | Download (5,033) | ||
| YY1 | BH-LCLs | GEO | Homo sapiens | GSE98477 | Download (4,062) | ||
| YY1 | HEP10-01008-LCLs | YY1mut | GEO | Homo sapiens | GSE98477 | Download (3,096) | |
| YY1 | HEP14-00120-LCLs | YY1mut | GEO | Homo sapiens | GSE98477 | Download (1,896) | |
| YY1 | HEP14-0079-LCLs | YY1mut | GEO | Homo sapiens | GSE98477 | Download (1,513) | |
| YY1 | HEP14-0080-LCLs | YY1mut | GEO | Homo sapiens | GSE98477 | Download (1,767) | |
| YY1 | JD-LCLs | GEO | Homo sapiens | GSE98477 | Download (2,911) | ||
| YY1 | JL-LCLs | GEO | Homo sapiens | GSE98477 | Download (2,903) | ||
| YY1 | PK-LCLs | GEO | Homo sapiens | GSE98477 | Download (6,014) | ||
| YY1 | WA01 | GEO | Homo sapiens | GSE39096 | Download (11,125) | ||
| YY2 | HeLa | GEO | Homo sapiens | GSE76856 | Download (6,040) | ||
| YY2 | HEK293 | ENCODE | Homo sapiens | ENCSR692HSE | Download (14,661) | ||
| ZAT10 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,436) |
| ZAT6 | Col-0 | Col-0_seedling | 3d-ABA | GEO | Arabidopsis thaliana | GSE80564 | Download (22,327) |
| ZAT6 | Col-0 | Col-0_seedling | 3d-EtOH | GEO | Arabidopsis thaliana | GSE80564 | Download (20,509) |
| ZBED1 | GM12878 | ENCODE | Homo sapiens | ENCSR207PFI | Download (15,681) | ||
| ZBED1 | K-562 | ENCODE | Homo sapiens | ENCSR286PCG | Download (14,321) | ||
| ZBTB1 | HEK293 | ENCODE | Homo sapiens | ENCSR927UJQ | Download (3,426) | ||
| ZBTB1 | MCF-7 | ENCODE | Homo sapiens | ENCSR309ELI | Download (2,922) | ||
| ZBTB10 | HEK293 | ENCODE | Homo sapiens | ENCSR004PLU | Download (20,399) | ||
| ZBTB11 | HEK293 | ENCODE | Homo sapiens | ENCSR882ZTS | Download (21,707) | ||
| ZBTB11 | K-562 | ENCODE | Homo sapiens | ENCSR331GDC | Download (6,889) | ||
| ZBTB11 | K-562 | ENCODE | Homo sapiens | ENCSR706BJO | Download (10,723) | ||
| ZBTB11 | MCF-7 | ENCODE | Homo sapiens | ENCSR155VDK | Download (2,676) | ||
| ZBTB12 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (288) | ||
| ZBTB12 | HEK293 | ENCODE | Homo sapiens | ENCSR543KOA | Download (11,148) | ||
| ZBTB14 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (14,947) | ||
| ZBTB16 | hESC | GEO | Homo sapiens | GSE75115 | Download (1,015) | ||
| ZBTB16 | KG-1 | GEO | Homo sapiens | GSE109619 | Download (2,629) | ||
| ZBTB16 | KG-1 | shEZH2 | GEO | Homo sapiens | GSE109619 | Download (13,785) | |
| ZBTB17 | HEK293 | ENCODE | Homo sapiens | ENCSR631WAA | Download (122,173) | ||
| ZBTB18 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (2,078) | ||
| ZBTB2 | K-562 | ENCODE | Homo sapiens | ENCSR230PTV | Download (28,102) | ||
| ZBTB2 | GM12878 | GEO | Homo sapiens | GSE97661 | Download (69,326) | ||
| ZBTB2 | Hep-G2 | GEO | Homo sapiens | GSE97661 | Download (46,439) | ||
| ZBTB20 | HEK293 | ENCODE | Homo sapiens | ENCSR460MBI | Download (40,383) | ||
| ZBTB21 | HEK293 | ENCODE | Homo sapiens | ENCSR321MSF | Download (23,674) | ||
| ZBTB24 | HCT-116 | GEO | Homo sapiens | GSE111683 | Download (27,469) | ||
| ZBTB26 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (15,867) | ||
| ZBTB26 | HEK293 | ENCODE | Homo sapiens | ENCSR229DYF | Download (20,335) | ||
| ZBTB26 | Hep-G2 | ENCODE | Homo sapiens | ENCSR184SVO | Download (158,647) | ||
| ZBTB33 | GM12878 | ENCODE | Homo sapiens | ENCSR000BHC | Download (6,185) | ||
| ZBTB33 | GM12878 | ENCODE | Homo sapiens | ENCSR542FLV | Download (13,753) | ||
| ZBTB33 | liver | ENCODE | Homo sapiens | ENCSR345YWJ | Download (4,595) | ||
| ZBTB33 | liver | ENCODE | Homo sapiens | ENCSR516HUP | Download (26,899) | ||
| ZBTB33 | A-549 | ENCODE | Homo sapiens | ENCSR000BPZ | Download (13,216) | ||
| ZBTB33 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BHR | Download (1,988) | ||
| ZBTB33 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BNA | Download (2,496) | ||
| ZBTB33 | K-562 | ENCODE | Homo sapiens | ENCSR000BKF | Download (2,247) | ||
| ZBTB33 | K-562 | ENCODE | Homo sapiens | ENCSR876GXA | Download (78,538) | ||
| ZBTB33 | MCF-7 | ENCODE | Homo sapiens | ENCSR231YFE | Download (10,958) | ||
| ZBTB33 | SK-N-SH | ENCODE | Homo sapiens | ENCSR000BTS | Download (9,043) | ||
| ZBTB33 | HCT-116 | ENCODE | Homo sapiens | ENCSR000BNY | Download (6,834) | ||
| ZBTB40 | MCF-7 | ENCODE | Homo sapiens | ENCSR318LVG | Download (13,024) | ||
| ZBTB40 | GM12878 | ENCODE | Homo sapiens | ENCSR189YYK | Download (23,187) | ||
| ZBTB40 | Hep-G2 | ENCODE | Homo sapiens | ENCSR525YFS | Download (6,804) | ||
| ZBTB40 | K-562 | ENCODE | Homo sapiens | ENCSR158RYZ | Download (6,322) | ||
| ZBTB40 | K-562 | ENCODE | Homo sapiens | ENCSR237VLT | Download (68,618) | ||
| ZBTB42 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (31,435) | ||
| ZBTB44 | HEK293 | ENCODE | Homo sapiens | ENCSR076STQ | Download (34,539) | ||
| ZBTB48 | HEK293 | ENCODE | Homo sapiens | ENCSR781EQJ | Download (39,054) | ||
| ZBTB48 | HeLa | GEO | Homo sapiens | GSE96776 | Download (202,922) | ||
| ZBTB48 | HeLa | ZBTB48-KO | GEO | Homo sapiens | GSE96776 | Download (271,463) | |
| ZBTB48 | U2OS | GEO | Homo sapiens | GSE96776 | Download (32,575) | ||
| ZBTB48 | U2OS | ZBTB48-KO | GEO | Homo sapiens | GSE96776 | Download (65,776) | |
| ZBTB48 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (120,329) | ||
| ZBTB49 | HEK293 | ENCODE | Homo sapiens | ENCSR924GRG | Download (245) | ||
| ZBTB5 | K-562 | ENCODE | Homo sapiens | ENCSR389PWB | Download (6,822) | ||
| ZBTB5 | K-562 | ENCODE | Homo sapiens | ENCSR786OQY | Download (3,394) | ||
| ZBTB6 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (28,415) | ||
| ZBTB6 | HEK293 | ENCODE | Homo sapiens | ENCSR619OUC | Download (13,497) | ||
| ZBTB7A | HEK293 | ENCODE | Homo sapiens | ENCSR773REP | Download (14,136) | ||
| ZBTB7A | Ishikawa | ENCODE | Homo sapiens | ENCSR000BSZ | Download (36,626) | ||
| ZBTB7A | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BQA | Download (33,540) | ||
| ZBTB7A | HUDEP-2 | GEO | Homo sapiens | GSE103445 | Download (40,042) | ||
| ZBTB7A | K-562 | ENCODE | Homo sapiens | ENCSR000BME | Download (66,716) | ||
| ZBTB7A | K-562 | GEO | Homo sapiens | GSE103445 | Download (61,938) | ||
| ZBTB7B | MCF-7 | ENCODE | Homo sapiens | ENCSR277BXW | Download (13,245) | ||
| ZBTB8A | HEK293 | ENCODE | Homo sapiens | ENCSR481FEC | Download (41,396) | ||
| ZC3H11A | A-549 | ENCODE | Homo sapiens | ENCSR277OOQ | Download (5,055) | ||
| ZC3H11A | K-562 | ENCODE | Homo sapiens | ENCSR000EFR | Download (2,725) | ||
| ZC3H8 | HCT-116 | GEO | Homo sapiens | GSE47938 | Download (11,226) | ||
| ZEB1 | MIA-PaCa-2 | GEO | Homo sapiens | GSE88734 | Download (4,028) | ||
| ZEB1 | MIA-PaCa-2 | WT | GEO | Homo sapiens | GSE88734 | Download (1,133) | |
| ZEB1 | NCI-H1975 | GEO | Homo sapiens | GSE106896 | Download (4,068) | ||
| ZEB1 | NCI-H1975 | resistant | GEO | Homo sapiens | GSE106896 | Download (11,177) | |
| ZEB1 | neuron | bipolar_doxy_4d | ENCODE | Homo sapiens | ENCSR418KUS | Download (19,957) | |
| ZEB1 | PDAC | GEO | Homo sapiens | GSE64557 | Download (19,983) | ||
| ZEB1 | RKO | GEO | Homo sapiens | GSE88734 | Download (10,422) | ||
| ZEB1 | GM12878 | ENCODE | Homo sapiens | ENCSR000BND | Download (50,700) | ||
| ZEB1 | HEK293 | ENCODE | Homo sapiens | ENCSR909HMT | Download (22,309) | ||
| ZEB1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR000BVN | Download (13,176) | ||
| ZEB2 | HEK293 | ENCODE | Homo sapiens | ENCSR417VWF | Download (61,847) | ||
| ZEB2 | liver | GEO | Homo sapiens | GSE103048 | Download (709) | ||
| ZEB2 | K-562 | ENCODE | Homo sapiens | ENCSR004GKA | Download (43,242) | ||
| ZEB2 | K-562 | ENCODE | Homo sapiens | ENCSR322CFO | Download (35,322) | ||
| ZFHX2 | HEK293 | ENCODE | Homo sapiens | ENCSR632SIM | Download (92,389) | ||
| ZFP1 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (7) | ||
| ZFP14 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (567) | ||
| ZFP28 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (3,773) | ||
| ZFP3 | HEK293 | ENCODE | Homo sapiens | ENCSR134QIE | Download (2,756) | ||
| ZFP36 | GM12878 | ENCODE | Homo sapiens | ENCSR900XDB | Download (31,075) | ||
| ZFP36 | A-549 | ENCODE | Homo sapiens | ENCSR294JWV | Download (10,181) | ||
| ZFP36 | Hep-G2 | ENCODE | Homo sapiens | ENCSR382XLA | Download (10,821) | ||
| ZFP36 | K-562 | ENCODE | Homo sapiens | ENCSR776CYN | Download (73,984) | ||
| ZFP36 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR184MFH | Download (13,784) | ||
| ZFP37 | HEK293 | ENCODE | Homo sapiens | ENCSR365GRX | Download (17,727) | ||
| ZFP41 | HEK293 | ENCODE | Homo sapiens | ENCSR348ONY | Download (185) | ||
| ZFP42 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (4,062) | ||
| ZFP64 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (50,984) | ||
| ZFP64 | HEK293 | ENCODE | Homo sapiens | ENCSR592KSP | Download (6,010) | ||
| ZFP64 | Hep-G2 | ENCODE | Homo sapiens | ENCSR487CPI | Download (55,130) | ||
| ZFP69 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (790) | ||
| ZFP69B | HEK293T | GEO | Homo sapiens | GSE78099 | Download (15,714) | ||
| ZFP69B | HEK293 | ENCODE | Homo sapiens | ENCSR381VYR | Download (35,280) | ||
| ZFP82 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (588) | ||
| ZFP90 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (119) | ||
| ZFP91 | K-562 | ENCODE | Homo sapiens | ENCSR898XMH | Download (10,886) | ||
| ZFX | K-562 | ENCODE | Homo sapiens | ENCSR920ASP | Download (19,077) | ||
| ZFX | MCF-7 | ENCODE | Homo sapiens | ENCSR435OQD | Download (10,011) | ||
| ZFX | MCF-7 | GEO | Homo sapiens | GSE102616 | Download (23,699) | ||
| ZFX | LNCaP-C4-2B | ENCODE | Homo sapiens | ENCSR027UFT | Download (6,793) | ||
| ZFX | HCT-116 | ENCODE | Homo sapiens | ENCSR503GVO | Download (12,608) | ||
| ZFX | HCT-116 | GEO | Homo sapiens | GSE102616 | Download (12,563) | ||
| ZFX | LNCaP-C4-2B | GEO | Homo sapiens | GSE102616 | Download (6,798) | ||
| ZFX | PrEC | GEO | Homo sapiens | GSE102616 | Download (9,067) | ||
| ZFX | DAOY | GEO | Homo sapiens | GSE45394 | Download (8,393) | ||
| ZFX | HEK293T | ENCODE | Homo sapiens | ENCSR218GSN | Download (308,783) | ||
| ZFX | HEK293T | GEO | Homo sapiens | GSE102616 | Download (507,568) | ||
| ZFX | NOMO1 | GEO | Homo sapiens | GSE43147 | Download (7,364) | ||
| ZFX | RPMI8402 | GEO | Homo sapiens | GSE43147 | Download (813) | ||
| ZGPAT | Hep-G2 | ENCODE | Homo sapiens | ENCSR704IGU | Download (84,281) | ||
| ZHD1 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (2,241) |
| ZHD14 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (812) |
| ZHD3 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (10,835) |
| ZHD5 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (1,880) |
| ZHD6 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (167) |
| ZHD6 | Col-0 | Col-0_leaves | tnt_colamp | GEO | Arabidopsis thaliana | GSE60141 | Download (16,787) |
| ZHD9 | Col-0 | Col-0_leaves | tnt_col | GEO | Arabidopsis thaliana | GSE60141 | Download (438) |
| ZHX1 | K-562 | ENCODE | Homo sapiens | ENCSR557RVF | Download (12,480) | ||
| ZHX1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR855WKQ | Download (46,395) | ||
| ZHX1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR887MXT | Download (25,254) | ||
| ZHX2 | MCF-7 | ENCODE | Homo sapiens | ENCSR876UYH | Download (11,387) | ||
| ZHX2 | Hep-G2 | ENCODE | Homo sapiens | ENCSR407BEZ | Download (26,523) | ||
| ZHX2 | 786-O | GEO | Homo sapiens | GSE109953 | Download (94,566) | ||
| ZIC2 | HEK293 | ENCODE | Homo sapiens | ENCSR728MWW | Download (87,817) | ||
| ZIC2 | BCBL-1 | latent | GEO | Homo sapiens | GSE102462 | Download (1,353) | |
| ZIC2 | BCBL-1 | lytic | GEO | Homo sapiens | GSE102462 | Download (351) | |
| ZIK1 | HEK293 | ENCODE | Homo sapiens | ENCSR074VAI | Download (1,082) | ||
| ZIK1 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (179) | ||
| ZIM2 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (21) | ||
| ZIM3 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (23,050) | ||
| ZIM3 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (26,117) | ||
| ZKSCAN1 | K-562 | ENCODE | Homo sapiens | ENCSR882ERE | Download (51,818) | ||
| ZKSCAN1 | MCF-7 | ENCODE | Homo sapiens | ENCSR449UFF | Download (10,731) | ||
| ZKSCAN1 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECJ | Download (8,604) | ||
| ZKSCAN1 | Hep-G2 | ENCODE | Homo sapiens | ENCSR157CAU | Download (29,522) | ||
| ZKSCAN2 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (4,824) | ||
| ZKSCAN3 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (79) | ||
| ZKSCAN5 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (6,334) | ||
| ZKSCAN8 | K-562 | ENCODE | Homo sapiens | ENCSR448UKK | Download (5,436) | ||
| ZMIZ1 | K-562 | ENCODE | Homo sapiens | ENCSR000EFQ | Download (19,295) | ||
| ZMIZ1 | K-562 | ENCODE | Homo sapiens | ENCSR907JPB | Download (1,511) | ||
| ZMYM2 | MCF-10A | GEO | Homo sapiens | GSE101921 | Download (1,013) | ||
| ZMYM3 | Hep-G2 | ENCODE | Homo sapiens | ENCSR848YWD | Download (20,738) | ||
| ZMYM3 | K-562 | ENCODE | Homo sapiens | ENCSR102KIN | Download (58,126) | ||
| ZMYM3 | GM12878 | GEO | Homo sapiens | GSE97661 | Download (15,398) | ||
| ZMYM3 | Hep-G2 | Ab_JH39-2-2F10 | GEO | Homo sapiens | GSE97661 | Download (24,921) | |
| ZMYM3 | Hep-G2 | AC_JH39-2-2B9 | GEO | Homo sapiens | GSE97661 | Download (14,974) | |
| ZMYM3 | Hep-G2 | AC_R259-2-2C3 | GEO | Homo sapiens | GSE97661 | Download (896) | |
| ZMYM3 | K-562 | Ab_JH39-2-2F10 | GEO | Homo sapiens | GSE97661 | Download (36,915) | |
| ZMYM3 | MCF-7 | GEO | Homo sapiens | GSE97661 | Download (1,554) | ||
| ZMYND11 | DU145 | ETS1KO | GEO | Homo sapiens | GSE86238 | Download (4,565) | |
| ZMYND11 | HeLa | GEO | Homo sapiens | GSE51672 | Download (16,998) | ||
| ZMYND11 | U2OS | SHZNYMD11 | GEO | Homo sapiens | GSE48423 | Download (192) | |
| ZMYND8 | HEK293 | GEO | Homo sapiens | GSE81696 | Download (1,833) | ||
| ZMYND8 | HEK293 | Flag-ZMYND8 | GEO | Homo sapiens | GSE81696 | Download (2,613) | |
| ZNF10 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (2,750) | ||
| ZNF10 | HEK293 | ENCODE | Homo sapiens | ENCSR019WUS | Download (23,697) | ||
| ZNF101 | HEK293 | ENCODE | Homo sapiens | ENCSR462FWS | Download (14,476) | ||
| ZNF101 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,493) | ||
| ZNF112 | HEK293 | ENCODE | Homo sapiens | ENCSR751PNN | Download (976) | ||
| ZNF114 | HEK293 | ENCODE | Homo sapiens | ENCSR985MYI | Download (312) | ||
| ZNF114 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (81) | ||
| ZNF12 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (676) | ||
| ZNF121 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (27,657) | ||
| ZNF121 | HEK293 | ENCODE | Homo sapiens | ENCSR224QDY | Download (35,548) | ||
| ZNF124 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (218) | ||
| ZNF132 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (185) | ||
| ZNF132 | HEK293 | ENCODE | Homo sapiens | ENCSR138RCE | Download (122) | ||
| ZNF133 | HEK293 | ENCODE | Homo sapiens | ENCSR283MWQ | Download (9,186) | ||
| ZNF133 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (8,793) | ||
| ZNF134 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (6,156) | ||
| ZNF135 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (335) | ||
| ZNF136 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (2,289) | ||
| ZNF138 | HEK293 | ENCODE | Homo sapiens | ENCSR551KAP | Download (415) | ||
| ZNF140 | HEK293 | ENCODE | Homo sapiens | ENCSR464KFG | Download (5,874) | ||
| ZNF140 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (4,702) | ||
| ZNF141 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (4,623) | ||
| ZNF143 | WA01 | ENCODE | Homo sapiens | ENCSR000EBW | Download (51,446) | ||
| ZNF143 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000ECO | Download (9,504) | ||
| ZNF143 | CUTLL1 | GEO | Homo sapiens | GSE29600 | Download (7,513) | ||
| ZNF143 | FLP143HA | T0 | GEO | Homo sapiens | GSE39263 | Download (47,904) | |
| ZNF143 | FLP143HA | T4 | GEO | Homo sapiens | GSE39263 | Download (64,426) | |
| ZNF143 | FLP76 | T4 | GEO | Homo sapiens | GSE39263 | Download (76,630) | |
| ZNF143 | HEK293T | GEO | Homo sapiens | GSE39263 | Download (3,709) | ||
| ZNF143 | HeLa | GEO | Homo sapiens | GSE31417 | Download (1,505) | ||
| ZNF143 | HeLa | GEO | Homo sapiens | GSE39263 | Download (19,925) | ||
| ZNF143 | GM12878 | ENCODE | Homo sapiens | ENCSR000DZL | Download (14,852) | ||
| ZNF143 | GM12878 | ENCODE | Homo sapiens | ENCSR936XTK | Download (45,536) | ||
| ZNF143 | WA09 | GEO | Homo sapiens | GSE105028 | Download (5,707) | ||
| ZNF143 | WA09 | heat-shock | GEO | Homo sapiens | GSE105028 | Download (8,043) | |
| ZNF143 | Hep-G2 | ENCODE | Homo sapiens | ENCSR101FJT | Download (44,439) | ||
| ZNF143 | K-562 | ENCODE | Homo sapiens | ENCSR000EGP | Download (37,535) | ||
| ZNF143 | HPBALL | GEO | Homo sapiens | GSE39263 | Download (1,905) | ||
| ZNF143 | K-562 | GEO | Homo sapiens | GSE39263 | Download (19,919) | ||
| ZNF143 | MCF-7 | GEO | Homo sapiens | GSE76454 | Download (63,978) | ||
| ZNF143 | MCF-7 | E2 | GEO | Homo sapiens | GSE76454 | Download (54,074) | |
| ZNF146 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (6,558) | ||
| ZNF146 | HEK293 | ENCODE | Homo sapiens | ENCSR689YFA | Download (20,991) | ||
| ZNF148 | HEK293 | ENCODE | Homo sapiens | ENCSR736ZKL | Download (2,714) | ||
| ZNF148 | K-562 | ENCODE | Homo sapiens | ENCSR018MSO | Download (17,958) | ||
| ZNF154 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (53) | ||
| ZNF155 | HEK293 | ENCODE | Homo sapiens | ENCSR801BWR | Download (428) | ||
| ZNF157 | HEK293 | ENCODE | Homo sapiens | ENCSR564YYW | Download (4,197) | ||
| ZNF157 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,054) | ||
| ZNF16 | HEK293 | ENCODE | Homo sapiens | ENCSR093HXE | Download (1,518) | ||
| ZNF165 | WHIM12 | GEO | Homo sapiens | GSE65937 | Download (145) | ||
| ZNF169 | HEK293 | ENCODE | Homo sapiens | ENCSR661AXW | Download (2,576) | ||
| ZNF17 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (957) | ||
| ZNF174 | HEK293 | ENCODE | Homo sapiens | ENCSR516VQK | Download (225) | ||
| ZNF175 | HEK293 | ENCODE | Homo sapiens | ENCSR103GAK | Download (398) | ||
| ZNF175 | Hep-G2 | ENCODE | Homo sapiens | ENCSR014RHK | Download (54,516) | ||
| ZNF175 | K-562 | ENCODE | Homo sapiens | ENCSR011PEI | Download (11,365) | ||
| ZNF175 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (9,930) | ||
| ZNF18 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (9,243) | ||
| ZNF18 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (152) | ||
| ZNF18 | GM12878 | GEO | Homo sapiens | GSE97661 | Download (8,101) | ||
| ZNF18 | K-562 | GEO | Homo sapiens | GSE97661 | Download (10,767) | ||
| ZNF18 | HEK293 | ENCODE | Homo sapiens | ENCSR977HTH | Download (22,287) | ||
| ZNF180 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,368) | ||
| ZNF181 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (14) | ||
| ZNF182 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (13,658) | ||
| ZNF184 | K-562 | ENCODE | Homo sapiens | ENCSR621ATC | Download (17,203) | ||
| ZNF184 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (320) | ||
| ZNF184 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (200) | ||
| ZNF184 | HEK293 | ENCODE | Homo sapiens | ENCSR020UPN | Download (21,499) | ||
| ZNF184 | K-562 | ENCODE | Homo sapiens | ENCSR546IHU | Download (9,870) | ||
| ZNF189 | HEK293 | ENCODE | Homo sapiens | ENCSR163RYW | Download (53,395) | ||
| ZNF189 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (4,942) | ||
| ZNF19 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (20,706) | ||
| ZNF19 | HEK293 | ENCODE | Homo sapiens | ENCSR811JJM | Download (513) | ||
| ZNF195 | HEK293 | ENCODE | Homo sapiens | ENCSR534NZI | Download (534) | ||
| ZNF197 | K-562 | ENCODE | Homo sapiens | ENCSR580IAO | Download (759) | ||
| ZNF197 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (415) | ||
| ZNF2 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (2,914) | ||
| ZNF2 | HEK293 | ENCODE | Homo sapiens | ENCSR011CKE | Download (44,497) | ||
| ZNF202 | HEK293 | ENCODE | Homo sapiens | ENCSR996FYT | Download (3,365) | ||
| ZNF202 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (13,654) | ||
| ZNF205 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (29,960) | ||
| ZNF207 | MCF-7 | ENCODE | Homo sapiens | ENCSR096KWU | Download (6,372) | ||
| ZNF207 | GM12878 | ENCODE | Homo sapiens | ENCSR117KWH | Download (21,785) | ||
| ZNF207 | Hep-G2 | ENCODE | Homo sapiens | ENCSR683CSF | Download (150) | ||
| ZNF207 | WA09 | GEO | Homo sapiens | GSE118632 | Download (8,946) | ||
| ZNF211 | HEK293 | ENCODE | Homo sapiens | ENCSR365DQH | Download (1,141) | ||
| ZNF212 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (52) | ||
| ZNF213 | HEK293 | ENCODE | Homo sapiens | ENCSR295GRB | Download (18,331) | ||
| ZNF214 | HEK293 | ENCODE | Homo sapiens | ENCSR993DED | Download (589) | ||
| ZNF214 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (133) | ||
| ZNF214 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (18) | ||
| ZNF217 | MCF-7 | ENCODE | Homo sapiens | ENCSR000EWU | Download (9,088) | ||
| ZNF217 | MCF-7 | ENCODE | Homo sapiens | ENCSR465XQW | Download (36,060) | ||
| ZNF217 | MCF-7 | ENCODE | Homo sapiens | ENCSR912ATU | Download (1,365) | ||
| ZNF217 | GM12878 | ENCODE | Homo sapiens | ENCSR764CZW | Download (21,715) | ||
| ZNF22 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (29,681) | ||
| ZNF221 | HEK293 | ENCODE | Homo sapiens | ENCSR194BBY | Download (577) | ||
| ZNF222 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,374) | ||
| ZNF223 | HEK293 | ENCODE | Homo sapiens | ENCSR906PCS | Download (3,576) | ||
| ZNF223 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (294) | ||
| ZNF224 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (648) | ||
| ZNF224 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (382) | ||
| ZNF225 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (93) | ||
| ZNF23 | HEK293 | ENCODE | Homo sapiens | ENCSR679KXF | Download (595) | ||
| ZNF235 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (5) | ||
| ZNF239 | HEK293 | ENCODE | Homo sapiens | ENCSR440COG | Download (3,571) | ||
| ZNF24 | GM12878 | ENCODE | Homo sapiens | ENCSR072PWP | Download (17,619) | ||
| ZNF24 | HEK293 | ENCODE | Homo sapiens | ENCSR984MDV | Download (29,346) | ||
| ZNF24 | Hep-G2 | ENCODE | Homo sapiens | ENCSR362NWP | Download (17,872) | ||
| ZNF24 | Hep-G2 | ENCODE | Homo sapiens | ENCSR791AGT | Download (797) | ||
| ZNF24 | K-562 | ENCODE | Homo sapiens | ENCSR099NCH | Download (43,737) | ||
| ZNF24 | K-562 | ENCODE | Homo sapiens | ENCSR117WTM | Download (22,178) | ||
| ZNF24 | K-562 | ENCODE | Homo sapiens | ENCSR385AHH | Download (17,385) | ||
| ZNF24 | K-562 | ENCODE | Homo sapiens | ENCSR695EQB | Download (50,858) | ||
| ZNF24 | MCF-7 | ENCODE | Homo sapiens | ENCSR503VTG | Download (22,544) | ||
| ZNF248 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,192) | ||
| ZNF248 | HEK293 | ENCODE | Homo sapiens | ENCSR338SEM | Download (1,248) | ||
| ZNF25 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (812) | ||
| ZNF250 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (360) | ||
| ZNF254 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (39) | ||
| ZNF257 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (1,724) | ||
| ZNF257 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (31,991) | ||
| ZNF26 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (4,497) | ||
| ZNF26 | HEK293 | ENCODE | Homo sapiens | ENCSR028EGI | Download (1,988) | ||
| ZNF260 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (3,608) | ||
| ZNF263 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (6,405) | ||
| ZNF263 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (14,846) | ||
| ZNF263 | HEK293 | ENCODE | Homo sapiens | ENCSR000EVD | Download (108,504) | ||
| ZNF263 | K-562 | ENCODE | Homo sapiens | ENCSR000EWN | Download (79,741) | ||
| ZNF264 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (8,249) | ||
| ZNF264 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (93) | ||
| ZNF266 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,096) | ||
| ZNF266 | HEK293 | ENCODE | Homo sapiens | ENCSR466VYP | Download (3,113) | ||
| ZNF267 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (294) | ||
| ZNF273 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (12,381) | ||
| ZNF274 | NT2-D1 | ENCODE | Homo sapiens | ENCSR000EWY | Download (597) | ||
| ZNF274 | WA01 | ENCODE | Homo sapiens | ENCSR000EUN | Download (769) | ||
| ZNF274 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000EVG | Download (147) | ||
| ZNF274 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (431) | ||
| ZNF274 | GM08714 | ENCODE | Homo sapiens | ENCSR000EUI | Download (944) | ||
| ZNF274 | GM12878 | ENCODE | Homo sapiens | ENCSR000EUK | Download (273) | ||
| ZNF274 | HEK293 | ENCODE | Homo sapiens | ENCSR178QVJ | Download (1,508) | ||
| ZNF274 | K-562 | ENCODE | Homo sapiens | ENCSR000EVX | Download (3,221) | ||
| ZNF28 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (11,427) | ||
| ZNF280A | HEK293 | GEO | Homo sapiens | GSE76494 | Download (14,325) | ||
| ZNF280A | K-562 | ENCODE | Homo sapiens | ENCSR370NFS | Download (4,662) | ||
| ZNF280C | HEK293 | ENCODE | Homo sapiens | ENCSR076KZW | Download (520) | ||
| ZNF280D | HEK293 | ENCODE | Homo sapiens | ENCSR451CYX | Download (9,789) | ||
| ZNF281 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (4,921) | ||
| ZNF282 | K-562 | ENCODE | Homo sapiens | ENCSR742TMU | Download (8,956) | ||
| ZNF282 | Hep-G2 | ENCODE | Homo sapiens | ENCSR752NDX | Download (1,742) | ||
| ZNF283 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (36,056) | ||
| ZNF284 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (303) | ||
| ZNF285 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (924) | ||
| ZNF287 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,045) | ||
| ZNF292 | HEK293 | ENCODE | Homo sapiens | ENCSR228PGT | Download (711) | ||
| ZNF3 | Hep-G2 | ENCODE | Homo sapiens | ENCSR182QWU | Download (37,307) | ||
| ZNF3 | K-562 | ENCODE | Homo sapiens | ENCSR195QFV | Download (65,020) | ||
| ZNF3 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (49) | ||
| ZNF30 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (15,808) | ||
| ZNF30 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (454) | ||
| ZNF300 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (31,236) | ||
| ZNF300 | HEK293 | ENCODE | Homo sapiens | ENCSR619QFT | Download (667) | ||
| ZNF302 | HEK293 | ENCODE | Homo sapiens | ENCSR513MGG | Download (453) | ||
| ZNF302 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (560) | ||
| ZNF304 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (710) | ||
| ZNF311 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (68) | ||
| ZNF311 | HEK293 | ENCODE | Homo sapiens | ENCSR096LSN | Download (1,937) | ||
| ZNF316 | K-562 | ENCODE | Homo sapiens | ENCSR167KBO | Download (127,640) | ||
| ZNF316 | K-562 | ENCODE | Homo sapiens | ENCSR200JYP | Download (23,353) | ||
| ZNF317 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (46,257) | ||
| ZNF317 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (5,090) | ||
| ZNF318 | K-562 | ENCODE | Homo sapiens | ENCSR334HSW | Download (2,438) | ||
| ZNF318 | K-562 | ENCODE | Homo sapiens | ENCSR352BJL | Download (6,107) | ||
| ZNF320 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (12,858) | ||
| ZNF320 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (2,866) | ||
| ZNF322 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (3,880) | ||
| ZNF324 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (17,249) | ||
| ZNF324 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (51) | ||
| ZNF324 | HEK293 | ENCODE | Homo sapiens | ENCSR768HOH | Download (20,705) | ||
| ZNF324B | HEK293T | GEO | Homo sapiens | GSE78099 | Download (4) | ||
| ZNF329 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (2,409) | ||
| ZNF331 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (3,722) | ||
| ZNF331 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (484) | ||
| ZNF333 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,228) | ||
| ZNF334 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (153,385) | ||
| ZNF335 | HEK293 | ENCODE | Homo sapiens | ENCSR328SUD | Download (59,541) | ||
| ZNF335 | Jurkat | GEO | Homo sapiens | GSE83116 | Download (1) | ||
| ZNF337 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,232) | ||
| ZNF33A | HEK293 | GEO | Homo sapiens | GSE76494 | Download (380) | ||
| ZNF33B | HEK293T | GEO | Homo sapiens | GSE78099 | Download (120) | ||
| ZNF34 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (537) | ||
| ZNF34 | HEK293 | ENCODE | Homo sapiens | ENCSR727PIC | Download (14,477) | ||
| ZNF341 | HEK293 | ENCODE | Homo sapiens | ENCSR185FOY | Download (39,711) | ||
| ZNF341 | LBCL | EBV-transformed | GEO | Homo sapiens | GSE107719 | Download (1,596) | |
| ZNF341 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (31,450) | ||
| ZNF341 | HIES | EBV-B_ZNF341_isoform1 | GEO | Homo sapiens | GSE113194 | Download (8,791) | |
| ZNF341 | HIES | EBV-B_ZNF341_isoform2 | GEO | Homo sapiens | GSE113194 | Download (10,125) | |
| ZNF341 | HIES | T-cells_anti-CD3_anti-CD28 | GEO | Homo sapiens | GSE113194 | Download (2,951) | |
| ZNF343 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (4,272) | ||
| ZNF35 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (59,722) | ||
| ZNF350 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (4,721) | ||
| ZNF350 | HEK293 | ENCODE | Homo sapiens | ENCSR854ORP | Download (4,481) | ||
| ZNF354A | HEK293 | GEO | Homo sapiens | GSE76494 | Download (628) | ||
| ZNF354A | HEK293T | GEO | Homo sapiens | GSE78099 | Download (377) | ||
| ZNF354B | K-562 | ENCODE | Homo sapiens | ENCSR674SCQ | Download (591) | ||
| ZNF354B | HEK293T | GEO | Homo sapiens | GSE78099 | Download (3) | ||
| ZNF354C | HEK293 | ENCODE | Homo sapiens | ENCSR289NSN | Download (376) | ||
| ZNF362 | HEK293 | ENCODE | Homo sapiens | ENCSR715QNO | Download (68,414) | ||
| ZNF366 | HEK293 | ENCODE | Homo sapiens | ENCSR106EBH | Download (77,728) | ||
| ZNF37A | HEK293 | ENCODE | Homo sapiens | ENCSR371LLY | Download (1,285) | ||
| ZNF37A | HEK293 | GEO | Homo sapiens | GSE76494 | Download (549) | ||
| ZNF383 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,420) | ||
| ZNF384 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (216,127) | ||
| ZNF384 | GM12878 | ENCODE | Homo sapiens | ENCSR000DYP | Download (4,698) | ||
| ZNF384 | HEK293T | ENCODE | Homo sapiens | ENCSR882ICT | Download (19,186) | ||
| ZNF384 | Hep-G2 | ENCODE | Homo sapiens | ENCSR101FJU | Download (46,078) | ||
| ZNF384 | K-562 | ENCODE | Homo sapiens | ENCSR000EFP | Download (39,288) | ||
| ZNF391 | HEK293 | ENCODE | Homo sapiens | ENCSR210MET | Download (19,720) | ||
| ZNF394 | HEK293 | ENCODE | Homo sapiens | ENCSR125DNC | Download (37,751) | ||
| ZNF394 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (753) | ||
| ZNF395 | K-562 | ENCODE | Homo sapiens | ENCSR462QZZ | Download (4,087) | ||
| ZNF398 | HEK293 | ENCODE | Homo sapiens | ENCSR676ZEF | Download (31,866) | ||
| ZNF398 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (149) | ||
| ZNF404 | HEK293 | ENCODE | Homo sapiens | ENCSR669GUM | Download (851) | ||
| ZNF407 | K-562 | ENCODE | Homo sapiens | ENCSR011NOZ | Download (4,087) | ||
| ZNF407 | K-562 | ENCODE | Homo sapiens | ENCSR439OCL | Download (2,560) | ||
| ZNF41 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (3,880) | ||
| ZNF410 | K-562 | GEO | Homo sapiens | GSE97661 | Download (5,883) | ||
| ZNF416 | HEK293 | ENCODE | Homo sapiens | ENCSR604CNJ | Download (659) | ||
| ZNF417 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (731) | ||
| ZNF418 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (611) | ||
| ZNF418 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (257) | ||
| ZNF423 | HEK293 | ENCODE | Homo sapiens | ENCSR477OJI | Download (10,261) | ||
| ZNF425 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (6,542) | ||
| ZNF426 | HEK293 | ENCODE | Homo sapiens | ENCSR224NFP | Download (9,586) | ||
| ZNF429 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (4,035) | ||
| ZNF430 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (141) | ||
| ZNF431 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (192) | ||
| ZNF432 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (200) | ||
| ZNF433 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (556) | ||
| ZNF433 | HEK293 | ENCODE | Homo sapiens | ENCSR586BRJ | Download (2,028) | ||
| ZNF436 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (4,804) | ||
| ZNF439 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (26) | ||
| ZNF44 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (2,739) | ||
| ZNF440 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (13,397) | ||
| ZNF441 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (4,664) | ||
| ZNF442 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (21) | ||
| ZNF443 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (29) | ||
| ZNF444 | MCF-7 | ENCODE | Homo sapiens | ENCSR038XIA | Download (40,656) | ||
| ZNF445 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (4,094) | ||
| ZNF449 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (7,369) | ||
| ZNF449 | HEK293 | ENCODE | Homo sapiens | ENCSR738SLS | Download (24,498) | ||
| ZNF45 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (46) | ||
| ZNF454 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (156) | ||
| ZNF454 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (128) | ||
| ZNF45A | MCF-7 | GEO | Homo sapiens | GSE97661 | Download (2,515) | ||
| ZNF460 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (561) | ||
| ZNF467 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (40,688) | ||
| ZNF468 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (6,618) | ||
| ZNF473 | HEK293 | ENCODE | Homo sapiens | ENCSR567XAM | Download (1,915) | ||
| ZNF479 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (37,606) | ||
| ZNF48 | HEK293 | ENCODE | Homo sapiens | ENCSR517DDU | Download (185) | ||
| ZNF48 | Hep-G2 | ENCODE | Homo sapiens | ENCSR415AEB | Download (121,410) | ||
| ZNF480 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (338) | ||
| ZNF483 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (498) | ||
| ZNF483 | BG01V | GEO | Homo sapiens | GSE97403 | Download (152) | ||
| ZNF484 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (295) | ||
| ZNF485 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (6,692) | ||
| ZNF486 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (13) | ||
| ZNF487 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,863) | ||
| ZNF488 | HEK293 | ENCODE | Homo sapiens | ENCSR363XBR | Download (1,972) | ||
| ZNF490 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (11,109) | ||
| ZNF491 | HEK293 | ENCODE | Homo sapiens | ENCSR897JCM | Download (225) | ||
| ZNF492 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (620) | ||
| ZNF493 | HEK293 | ENCODE | Homo sapiens | ENCSR550ZZK | Download (544) | ||
| ZNF496 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (17,587) | ||
| ZNF501 | HEK293 | ENCODE | Homo sapiens | ENCSR461ZJT | Download (16,983) | ||
| ZNF502 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (378) | ||
| ZNF506 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (5,649) | ||
| ZNF507 | K-562 | ENCODE | Homo sapiens | ENCSR598TIR | Download (1,802) | ||
| ZNF507 | MCF-7 | ENCODE | Homo sapiens | ENCSR419ODQ | Download (1,659) | ||
| ZNF510 | HEK293 | ENCODE | Homo sapiens | ENCSR595FAO | Download (3,464) | ||
| ZNF512 | K-562 | ENCODE | Homo sapiens | ENCSR591CCL | Download (14,048) | ||
| ZNF512B | MCF-7 | ENCODE | Homo sapiens | ENCSR555DCF | Download (7,142) | ||
| ZNF512B | MCF-7 | ENCODE | Homo sapiens | ENCSR761LRR | Download (15,138) | ||
| ZNF513 | HEK293 | ENCODE | Homo sapiens | ENCSR503DPC | Download (12,526) | ||
| ZNF514 | HEK293 | ENCODE | Homo sapiens | ENCSR897RVP | Download (410) | ||
| ZNF518A | HEK293 | ENCODE | Homo sapiens | ENCSR159GFL | Download (23,915) | ||
| ZNF519 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (14,287) | ||
| ZNF521 | HEK293 | ENCODE | Homo sapiens | ENCSR665KYH | Download (1,134) | ||
| ZNF524 | HEK293 | ENCODE | Homo sapiens | ENCSR418NZA | Download (15,247) | ||
| ZNF525 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (46) | ||
| ZNF527 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (88) | ||
| ZNF528 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (66,934) | ||
| ZNF528 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (716) | ||
| ZNF529 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,029) | ||
| ZNF529 | HEK293 | ENCODE | Homo sapiens | ENCSR754SOI | Download (1,792) | ||
| ZNF530 | HEK293 | ENCODE | Homo sapiens | ENCSR701RXW | Download (937) | ||
| ZNF530 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (659) | ||
| ZNF532 | NMC24335 | GEO | Homo sapiens | GSE96775 | Download (31,356) | ||
| ZNF534 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (42,246) | ||
| ZNF540 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (66) | ||
| ZNF543 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,044) | ||
| ZNF544 | HEK293 | ENCODE | Homo sapiens | ENCSR167XFW | Download (1,225) | ||
| ZNF547 | HEK293 | ENCODE | Homo sapiens | ENCSR909TSW | Download (4,458) | ||
| ZNF547 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (1,957) | ||
| ZNF547 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (3,153) | ||
| ZNF548 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (137) | ||
| ZNF548 | HEK293 | ENCODE | Homo sapiens | ENCSR892ZTO | Download (2,462) | ||
| ZNF549 | HEK293 | ENCODE | Homo sapiens | ENCSR185QFX | Download (6,235) | ||
| ZNF549 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (11,136) | ||
| ZNF550 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (23) | ||
| ZNF552 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (16) | ||
| ZNF554 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (40,080) | ||
| ZNF555 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (969) | ||
| ZNF555 | HEK293 | ENCODE | Homo sapiens | ENCSR767XSF | Download (6,825) | ||
| ZNF557 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,148) | ||
| ZNF558 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (3,957) | ||
| ZNF558 | HEK293 | ENCODE | Homo sapiens | ENCSR447ZTA | Download (16,546) | ||
| ZNF560 | HEK293 | ENCODE | Homo sapiens | ENCSR538RDA | Download (1,595) | ||
| ZNF561 | HEK293 | ENCODE | Homo sapiens | ENCSR125ULS | Download (33,638) | ||
| ZNF561 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (605) | ||
| ZNF562 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (33) | ||
| ZNF563 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (4,056) | ||
| ZNF564 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (36) | ||
| ZNF565 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (3,494) | ||
| ZNF566 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (241) | ||
| ZNF567 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (978) | ||
| ZNF57 | BG01V | GEO | Homo sapiens | GSE97403 | Download (248) | ||
| ZNF570 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (73) | ||
| ZNF571 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (458) | ||
| ZNF571 | HEK293 | ENCODE | Homo sapiens | ENCSR421QXA | Download (1,066) | ||
| ZNF573 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (18,556) | ||
| ZNF574 | MCF-7 | ENCODE | Homo sapiens | ENCSR402JAC | Download (1,473) | ||
| ZNF574 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (31,413) | ||
| ZNF577 | HEK293 | ENCODE | Homo sapiens | ENCSR776MDR | Download (14,417) | ||
| ZNF579 | MCF-7 | ENCODE | Homo sapiens | ENCSR018MQH | Download (18,775) | ||
| ZNF580 | HEK293 | ENCODE | Homo sapiens | ENCSR668HOP | Download (38,252) | ||
| ZNF580 | Hep-G2 | ENCODE | Homo sapiens | ENCSR173CTF | Download (99,460) | ||
| ZNF582 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (480) | ||
| ZNF582 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,410) | ||
| ZNF584 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (196) | ||
| ZNF584 | K-562 | ENCODE | Homo sapiens | ENCSR149ZBI | Download (3,021) | ||
| ZNF585A | HEK293T | GEO | Homo sapiens | GSE78099 | Download (6,617) | ||
| ZNF585B | HEK293 | ENCODE | Homo sapiens | ENCSR011XCI | Download (6,341) | ||
| ZNF586 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (1,957) | ||
| ZNF587 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (243) | ||
| ZNF589 | K-562 | ENCODE | Homo sapiens | ENCSR603XLW | Download (7,709) | ||
| ZNF592 | MCF-7 | ENCODE | Homo sapiens | ENCSR028NUR | Download (296) | ||
| ZNF592 | MCF-7 | ENCODE | Homo sapiens | ENCSR701AQS | Download (9,036) | ||
| ZNF592 | GM12878 | ENCODE | Homo sapiens | ENCSR173ZVL | Download (5,590) | ||
| ZNF592 | K-562 | ENCODE | Homo sapiens | ENCSR249BHQ | Download (39,515) | ||
| ZNF595 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (362) | ||
| ZNF596 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (1,550) | ||
| ZNF596 | HEK293 | ENCODE | Homo sapiens | ENCSR344SBD | Download (14,023) | ||
| ZNF597 | GM12878 | GEO | Homo sapiens | GSE97661 | Download (7,544) | ||
| ZNF600 | HEK293 | ENCODE | Homo sapiens | ENCSR017QBI | Download (152,273) | ||
| ZNF605 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (6,436) | ||
| ZNF610 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (2,701) | ||
| ZNF610 | HEK293 | ENCODE | Homo sapiens | ENCSR691TXI | Download (16,422) | ||
| ZNF611 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,258) | ||
| ZNF613 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (584) | ||
| ZNF614 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (102) | ||
| ZNF614 | Hep-G2 | ENCODE | Homo sapiens | ENCSR011CIR | Download (99,292) | ||
| ZNF615 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (24) | ||
| ZNF616 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (934) | ||
| ZNF619 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (48) | ||
| ZNF620 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (17) | ||
| ZNF621 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (94) | ||
| ZNF621 | HEK293 | ENCODE | Homo sapiens | ENCSR858FAN | Download (1,761) | ||
| ZNF622 | GM12878 | ENCODE | Homo sapiens | ENCSR075FNZ | Download (137) | ||
| ZNF623 | HEK293 | ENCODE | Homo sapiens | ENCSR022IZK | Download (15,519) | ||
| ZNF624 | HEK293 | ENCODE | Homo sapiens | ENCSR878DES | Download (738) | ||
| ZNF626 | HEK293 | ENCODE | Homo sapiens | ENCSR588MQZ | Download (4,245) | ||
| ZNF626 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (135) | ||
| ZNF627 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (72) | ||
| ZNF629 | HEK293 | ENCODE | Homo sapiens | ENCSR351NON | Download (59,278) | ||
| ZNF639 | HEK293 | ENCODE | Homo sapiens | ENCSR080CST | Download (25,487) | ||
| ZNF639 | K-562 | ENCODE | Homo sapiens | ENCSR497VFH | Download (5,818) | ||
| ZNF639 | K-562 | Ab_R270-2-1E7 | GEO | Homo sapiens | GSE97661 | Download (2,941) | |
| ZNF639 | K-562 | ENCODE | Homo sapiens | ENCSR845BCL | Download (9,590) | ||
| ZNF639 | K-562 | ENCODE | Homo sapiens | ENCSR949NVY | Download (18,293) | ||
| ZNF641 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (253) | ||
| ZNF644 | K-562 | ENCODE | Homo sapiens | ENCSR729HVR | Download (6,924) | ||
| ZNF644 | HEK293T | GEO | Homo sapiens | GSE62616 | Download (1,202) | ||
| ZNF644 | Hep-G2 | ENCODE | Homo sapiens | ENCSR578CXC | Download (81,164) | ||
| ZNF645 | HEK293 | ENCODE | Homo sapiens | ENCSR776LDJ | Download (2,691) | ||
| ZNF649 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,169) | ||
| ZNF652 | Hep-G2 | ENCODE | Homo sapiens | ENCSR502GAX | Download (60,117) | ||
| ZNF654 | HEK293 | ENCODE | Homo sapiens | ENCSR504VDV | Download (34,532) | ||
| ZNF658 | HEK293 | ENCODE | Homo sapiens | ENCSR762DQP | Download (533) | ||
| ZNF660 | HEK293 | ENCODE | Homo sapiens | ENCSR283DOU | Download (55,970) | ||
| ZNF662 | HEK293 | ENCODE | Homo sapiens | ENCSR483VJQ | Download (1,102) | ||
| ZNF662 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (457) | ||
| ZNF664 | HEK293 | ENCODE | Homo sapiens | ENCSR714LZQ | Download (33,808) | ||
| ZNF667 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (4,056) | ||
| ZNF667 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (38) | ||
| ZNF669 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (447) | ||
| ZNF669 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (71) | ||
| ZNF670 | HEK293 | ENCODE | Homo sapiens | ENCSR233MWH | Download (409) | ||
| ZNF671 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (3,279) | ||
| ZNF674 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (5,088) | ||
| ZNF675 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (2,574) | ||
| ZNF677 | HEK293 | ENCODE | Homo sapiens | ENCSR279KDC | Download (3,779) | ||
| ZNF680 | HEK293 | ENCODE | Homo sapiens | ENCSR307CKC | Download (9,323) | ||
| ZNF680 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (11,385) | ||
| ZNF681 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (217) | ||
| ZNF684 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (424) | ||
| ZNF687 | MCF-7 | ENCODE | Homo sapiens | ENCSR899BKM | Download (24,882) | ||
| ZNF687 | GM12878 | ENCODE | Homo sapiens | ENCSR859FDL | Download (26,977) | ||
| ZNF69 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (10) | ||
| ZNF692 | HEK293 | ENCODE | Homo sapiens | ENCSR418MKG | Download (55,199) | ||
| ZNF695 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (10,915) | ||
| ZNF697 | HEK293 | ENCODE | Homo sapiens | ENCSR217QCK | Download (7,980) | ||
| ZNF7 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (214) | ||
| ZNF701 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (720) | ||
| ZNF701 | HEK293 | ENCODE | Homo sapiens | ENCSR547TGL | Download (921) | ||
| ZNF704 | HEK293 | ENCODE | Homo sapiens | ENCSR194KWL | Download (600) | ||
| ZNF705G | HEK293T | GEO | Homo sapiens | GSE78099 | Download (4) | ||
| ZNF707 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,426) | ||
| ZNF707 | HEK293 | ENCODE | Homo sapiens | ENCSR854IPI | Download (1,923) | ||
| ZNF708 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (621) | ||
| ZNF708 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,027) | ||
| ZNF714 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (922) | ||
| ZNF716 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,511) | ||
| ZNF730 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,020) | ||
| ZNF736 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (36,769) | ||
| ZNF74 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (40) | ||
| ZNF740 | K-562 | ENCODE | Homo sapiens | ENCSR737UST | Download (3,471) | ||
| ZNF740 | K-562 | ENCODE | Homo sapiens | ENCSR532EMP | Download (1,284) | ||
| ZNF747 | HEK293 | ENCODE | Homo sapiens | ENCSR497IAS | Download (749) | ||
| ZNF749 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (120) | ||
| ZNF750 | keratinocyte | diff | GEO | Homo sapiens | GSE57702 | Download (55,297) | |
| ZNF75A | K-562 | GEO | Homo sapiens | GSE97661 | Download (3,492) | ||
| ZNF76 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (6,707) | ||
| ZNF76 | HEK293 | ENCODE | Homo sapiens | ENCSR072LQF | Download (20,111) | ||
| ZNF764 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (28) | ||
| ZNF765 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,462) | ||
| ZNF766 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (18,418) | ||
| ZNF766 | K-562 | ENCODE | Homo sapiens | ENCSR194IJN | Download (568) | ||
| ZNF768 | HEK293 | ENCODE | Homo sapiens | ENCSR070HWF | Download (4,946) | ||
| ZNF768 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (4,091) | ||
| ZNF77 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (257) | ||
| ZNF770 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (46,196) | ||
| ZNF770 | HEK293 | ENCODE | Homo sapiens | ENCSR242BGR | Download (77,725) | ||
| ZNF774 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (102) | ||
| ZNF776 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (245) | ||
| ZNF776 | HEK293 | ENCODE | Homo sapiens | ENCSR274LVQ | Download (2,716) | ||
| ZNF777 | HEK293 | ENCODE | Homo sapiens | ENCSR295BIP | Download (4,766) | ||
| ZNF777 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (5,887) | ||
| ZNF778 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (10,672) | ||
| ZNF778 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (6,639) | ||
| ZNF780A | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,120) | ||
| ZNF781 | HEK293 | ENCODE | Homo sapiens | ENCSR777YSB | Download (7,683) | ||
| ZNF782 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (43) | ||
| ZNF783 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (3,459) | ||
| ZNF784 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (869) | ||
| ZNF785 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (15) | ||
| ZNF785 | HEK293 | ENCODE | Homo sapiens | ENCSR950ACO | Download (7,049) | ||
| ZNF786 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (4,657) | ||
| ZNF789 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (401) | ||
| ZNF79 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (268) | ||
| ZNF790 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (38) | ||
| ZNF791 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (622) | ||
| ZNF791 | HEK293 | ENCODE | Homo sapiens | ENCSR775HFF | Download (4,307) | ||
| ZNF792 | HEK293 | ENCODE | Homo sapiens | ENCSR679STZ | Download (5,581) | ||
| ZNF792 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,995) | ||
| ZNF799 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (263) | ||
| ZNF8 | MCF-7 | ENCODE | Homo sapiens | ENCSR785UCD | Download (3,670) | ||
| ZNF8 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (4,396) | ||
| ZNF8 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (75) | ||
| ZNF805 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (35) | ||
| ZNF808 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,436) | ||
| ZNF81 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (197) | ||
| ZNF816 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (3,046) | ||
| ZNF816 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (132) | ||
| ZNF823 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (19,557) | ||
| ZNF83 | K-562 | ENCODE | Homo sapiens | ENCSR257XVY | Download (3,275) | ||
| ZNF830 | HEK293 | ENCODE | Homo sapiens | ENCSR781DPA | Download (319) | ||
| ZNF830 | K-562 | ENCODE | Homo sapiens | ENCSR033NQK | Download (2,956) | ||
| ZNF830 | K-562 | ENCODE | Homo sapiens | ENCSR851XLW | Download (895) | ||
| ZNF837 | HEK293 | ENCODE | Homo sapiens | ENCSR711UOA | Download (928) | ||
| ZNF84 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,781) | ||
| ZNF843 | HEK293 | ENCODE | Homo sapiens | ENCSR502KPJ | Download (30,755) | ||
| ZNF846 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (821) | ||
| ZNF85 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (1,605) | ||
| ZNF85 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (38) | ||
| ZNF860 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (300) | ||
| ZNF879 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (630) | ||
| ZNF880 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (117) | ||
| ZNF891 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (125) | ||
| ZNF90 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (21,141) | ||
| ZNF92 | retina | pigment | GEO | Homo sapiens | GSE60024 | Download (22,634) | |
| ZNF93 | HEK293T | GEO | Homo sapiens | GSE78099 | Download (1,931) | ||
| ZSCAN16 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (64,334) | ||
| ZSCAN16 | HEK293 | ENCODE | Homo sapiens | ENCSR864VJE | Download (5,590) | ||
| ZSCAN18 | HEK293 | ENCODE | Homo sapiens | ENCSR721QZV | Download (2,396) | ||
| ZSCAN2 | Hep-G2 | GEO | Homo sapiens | GSE97661 | Download (1,280) | ||
| ZSCAN2 | MCF-7 | GEO | Homo sapiens | GSE97661 | Download (1,638) | ||
| ZSCAN21 | HEK293 | ENCODE | Homo sapiens | ENCSR253CKN | Download (34,573) | ||
| ZSCAN22 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (38,454) | ||
| ZSCAN23 | HEK293 | ENCODE | Homo sapiens | ENCSR705ASR | Download (8,436) | ||
| ZSCAN26 | HEK293 | ENCODE | Homo sapiens | ENCSR501NSC | Download (742) | ||
| ZSCAN29 | GM12878 | ENCODE | Homo sapiens | ENCSR412YGM | Download (3,099) | ||
| ZSCAN29 | K-562 | ENCODE | Homo sapiens | ENCSR175SZH | Download (12,938) | ||
| ZSCAN29 | K-562 | ENCODE | Homo sapiens | ENCSR635EXI | Download (2,877) | ||
| ZSCAN29 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (347) | ||
| ZSCAN30 | HEK293 | ENCODE | Homo sapiens | ENCSR768VNZ | Download (30,273) | ||
| ZSCAN31 | HEK293 | GEO | Homo sapiens | GSE76494 | Download (1,970) | ||
| ZSCAN4 | HEK293 | ENCODE | Homo sapiens | ENCSR211GNP | Download (38,474) | ||
| ZSCAN5A | HEK293 | ENCODE | Homo sapiens | ENCSR357QJR | Download (7,631) | ||
| ZSCAN5A | BeWo | GEO | Homo sapiens | GSE85009 | Download (166,037) | ||
| ZSCAN5C | HEK293 | ENCODE | Homo sapiens | ENCSR731AGO | Download (24,753) | ||
| ZSCAN5D | BeWo | GEO | Homo sapiens | GSE85009 | Download (118,555) | ||
| ZXDB | HEK293 | ENCODE | Homo sapiens | ENCSR559IOZ | Download (43,653) | ||
| ZXDC | MCF-7 | GEO | Homo sapiens | GSE97661 | Download (21,626) | ||
| ZZZ3 | K-562 | ENCODE | Homo sapiens | ENCSR780BBJ | Download (13,014) | ||
| ZZZ3 | HeLa-S3 | ENCODE | Homo sapiens | ENCSR000DNT | Download (173) | ||
| ZZZ3 | GM12878 | ENCODE | Homo sapiens | ENCSR000DNQ | Download (130) | ||
| Target name | Ecotype | Biotype | Condition | Source | Species | Experiment | Peaks |